Method for improving activity of gene knockout and base editing system by using small molecule compound and application method thereof
A small molecule compound and gene knockout technology, applied in the field of genetic engineering, can solve the problems of low activity of precise site-specific gene editing, low efficiency of gene knockout and base editing system, etc., and achieve the effect of high gene editing efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
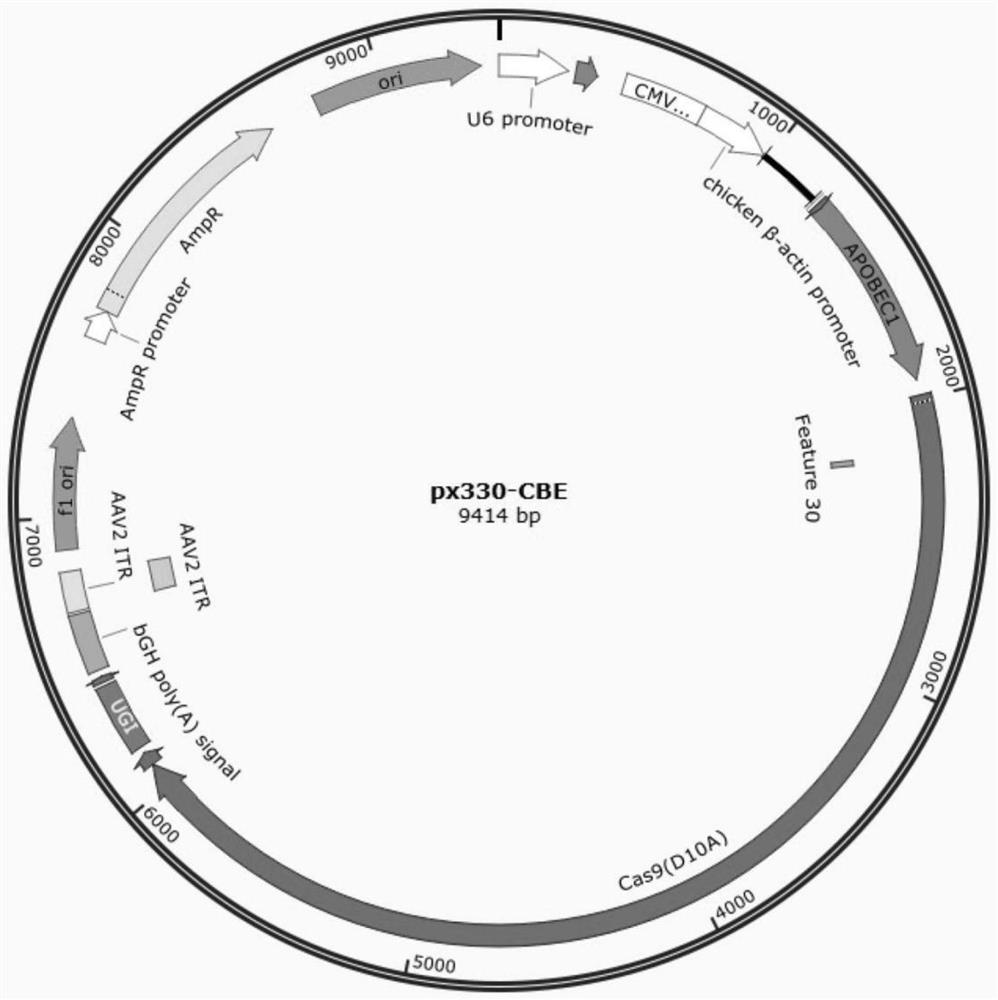
[0049] Embodiment 1: Construction of px330-CBE expression vector
[0050] (1) Experimental materials include:
[0051] Primers, restriction endonucleases, plasmid recombination kits, plasmid extraction kits, gel recovery kits, high-fidelity DNA polymerase, and genomic DNA extraction kits according to conventional methods. The plasmid pZHW-PBE contains the CBE sequence: SEQ ID NO.1. Plasmid px330, plasmid pCMV-ABEmax, plasmid pJET-U6. Plasmid pJET-U6 is the backbone of pJET1.2 (CloneJETPCR Cloning Kit, Thermo Fisher Scientific) connected with U6 and scaffold fragments (see SEQ ID NO.2 for the fragment sequence). Transfection Reagent Turbofect and T4 DNA Ligase.
[0052] Specifically, SEQ ID NO.2 U6-scaffold sequence
[0053] gagggcctatttcccatgattccttcatatttgcatatacgatacaaggctgttagagagataattggaattaatttgactgtaaacacaaagatattagtacaaaatacgtgacgtagaaagtaataatttcttgggtagtttgcagttttaaaattatgttttaaaatggactatcatatgcttaccgtaacttgaaagtatttcgatttcttggctttatatatcttgtggaaaggacgaaacaccgggt...
Embodiment 2
[0068] Embodiment 2: the establishment of the BFP reporter system that is used for CBE activity test
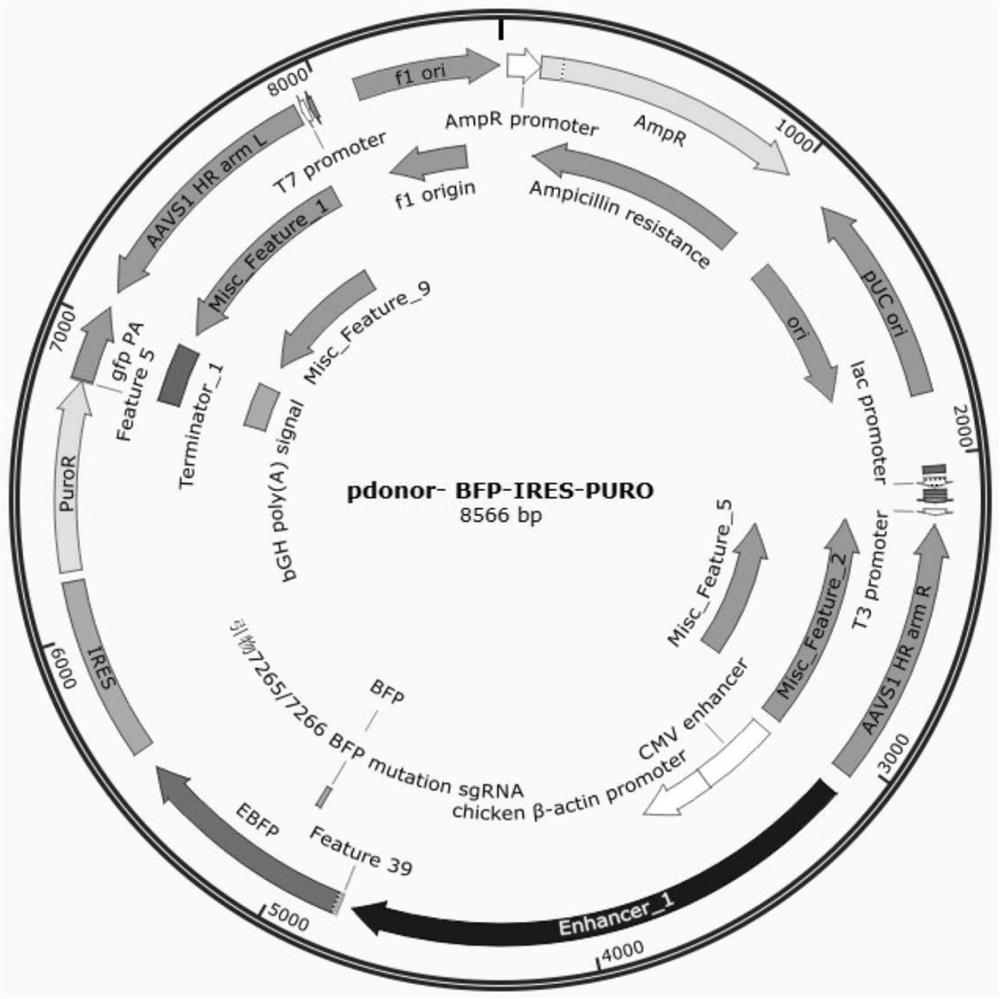
[0069] (1) Construct the px330-AAVS1 vector, which contains the sgRNA targeting the AAVS1 site (see Table 1 for the sequences AAVS1-sgRNA-F and AAVS1-sgRNA-R). Construct a vector (pdonor-BFP-IRES-PURO) expressing BFP with a homology arm of AAVS1 site, which contains AAVS1 homology arm, blue fluorescent protein (BFP), IRES and puromycin (Puromycin) resistance gene .
[0070] (2) Recovery of HEK-293 cells: Take out the tube of cryopreserved HEK-293 cells from liquid nitrogen, immediately put it into a 37°C water bath, shake slightly, and wait until the liquid is completely melted (about 1-1.5min), 1000rpm , and centrifuged for 3min; take it out and wipe it with 75% alcohol and put it on the ultra-clean workbench; discard the supernatant, add 1ml of cell culture medium to resuspend the cells, and then transfer to a 10cm culture dish containing 10ml of culture medium. Shake gen...
Embodiment 5
[0090] Example 5: Establishment of HEK293-ABCA4 cell line
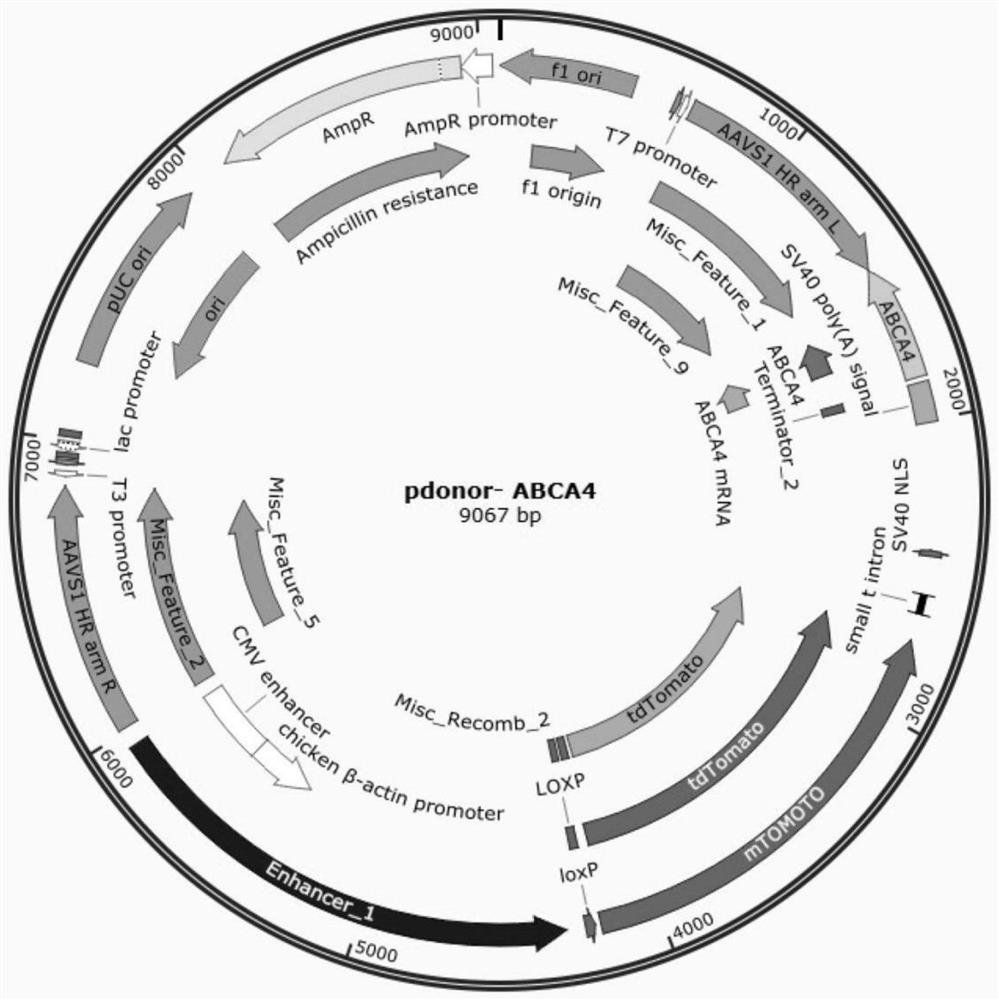
[0091] (1) Construction of recombinant vector pdonor-ABCA4:
[0092] The partial sequence of the mutant ABCA4 gene (as shown in 4) was amplified by PCR and cloned into the vector pdonor-BFP-IRES-PURO to obtain the recombinant pdonor-ABCA4 vector.
[0093] (2) Construction of a monoclonal HEK293 cell line integrated with the mutant ABCA4 gene:
[0094] Step 1: HEK293 cells were seeded in a 12-well plate after counting, and the cell density in each well was 1.6×10 5 cells / well;
[0095] Step 2: Cell transfection was carried out after 24 hours. The experimental group was co-transfected with px330-AAVS1 and pdonor-ABCA4 vectors, and the negative control group was co-transfected with px330 and pdonor-ABCA4 vectors. The rest of the conditions were the same as those of the experimental group;
[0096] Step 3: Change the medium 24 hours after transfection, and transfer to a 10cm dish after 48 hours;
[0097] Step 4: After...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com