Rice bacterial leaf blight resistant protein, and coding gene and application thereof
A technology for rice bacterial blight and bacterial blight resistance, which can be applied in application, genetic engineering, plant genetic improvement and other directions, and can solve the problems of inability to analyze and predict candidate genes.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] The isolation clone of embodiment 1Xa7 gene
[0029] On the basis of the previous research of the present invention, it was found that the genome sequence near the Xa7 gene locus is quite different from the common rice reference genomes Nipponbare, 9311, Minghui 63 and Zhenshan 97. Defining the genome sequence of this region is the prerequisite for the cloning of this gene. The present invention constructs the genome BAC library of IRBB7, an Xa7-resistant variety, screens the library with closely linked molecular markers on both sides of Xa7, catches positive clones, and sequences the inserts of the positive clones, thereby obtaining a complete and accurate genome sequence of the region.
[0030] The rice variety IRBB7 has been disclosed in the document "Ogawa et al., 1991, Breeding of near-isogenic lines of rice with single genes for resistance to bacterial blight pathogen Xanthomonas campestris pv. oryzae. Japan J Breed, 41:523-529.").
[0031] The plant material for...
Embodiment 2
[0041] The sequence structure and expression characteristics of embodiment 2Xa7 gene
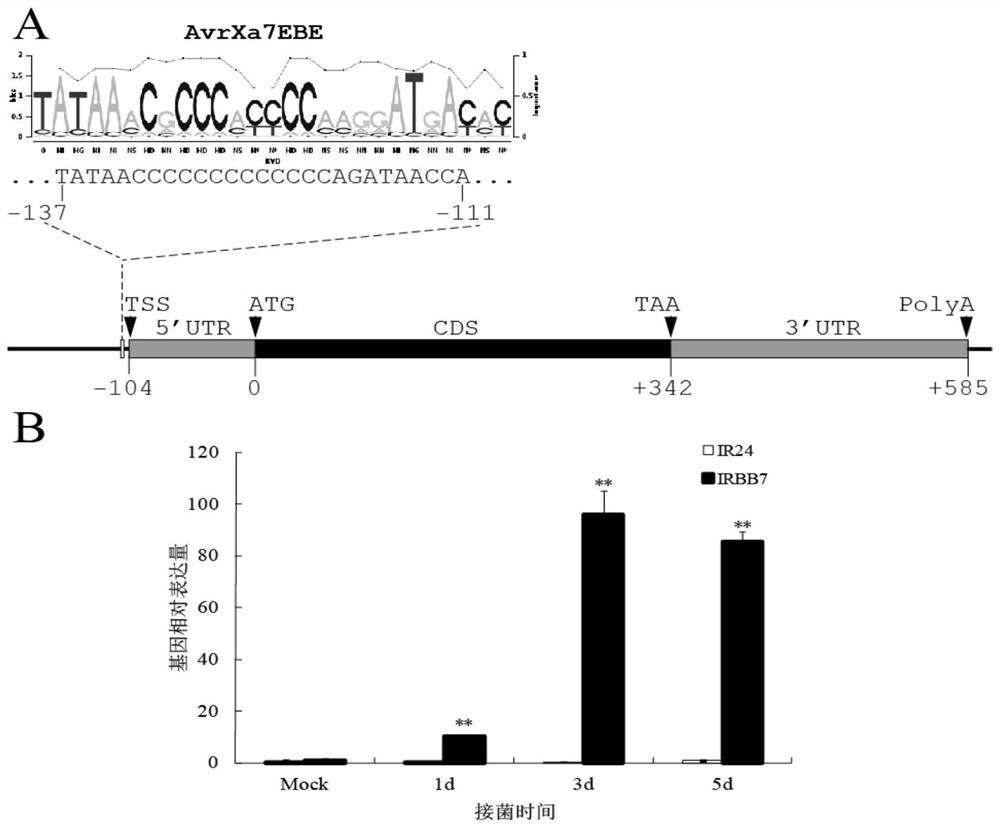
[0042] Sequence structure analysis of the Xa7 gene: total RNA was extracted from the near-isogenic line IRBB7 carrying the Xa7 gene, via Invitrogen TM GeneRacer TM Kit amplifies the full-length 5' and 3' ends of Xa7, respectively. Among them, the specific primer for 5'RACE amplification is 5'-TGCCACCGATGAGGTAATCCTGC-3', and the specific primer for 3'RACE amplification is 5'-CCTCCTCGGAATCTGGCTCATGTC-3'. RACE product passed pEASY TM -Blunt Zero CloningKit for cloning. After sequencing, it was determined that the transcription start site (TSS) of Xa7 was located at 104bp upstream of the initiation codon (ATG); at the same time, it was determined that the 3'UTR length of Xa7 was 253bp ( figure 2 A).
[0043] Analysis of the expression pattern of Xa7 gene: IRBB7 and IR24 were inoculated with Xanthobacterium sourbacillus (PXO86) at the booting stage by "leaf-cutting method" and samples we...
Embodiment 3
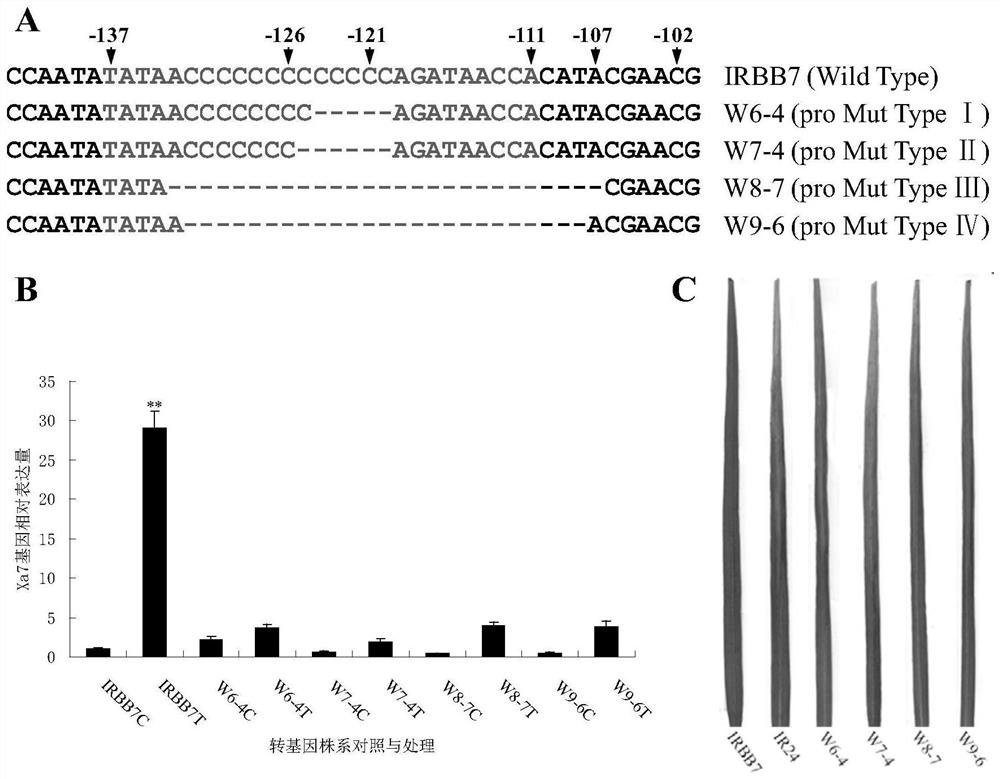
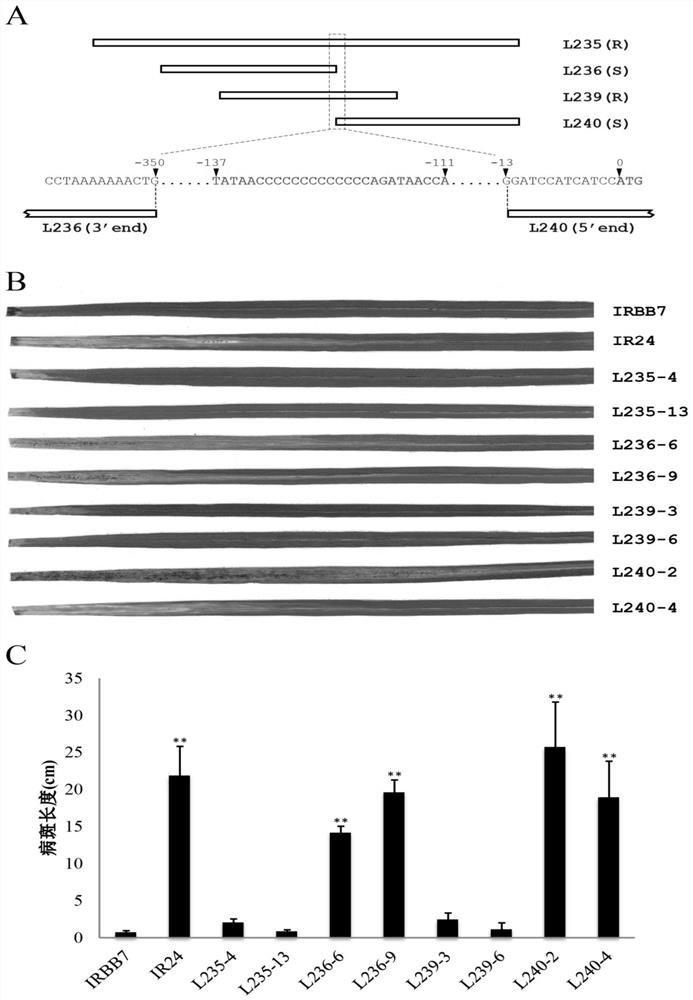
[0051] Example 3 The key functional sites of the Xa7 gene were verified by CRISPR / Cas9-mediated gene knockout
[0052] In order to further verify the functions of the Xa7 promoter region AvrXa7EBE and CDS region sequences, the present invention also utilizes the CRISPR / Cas9 system to construct gene knockout transgenic lines of these two functional regions. The vector used for gene knockout is the binary expression vector pYLCRISPR / Cas9P provided by Liu Yaoguang Laboratory of South China Agricultural University ubi -H (disclosed in the literature "Ma et al.2015, Arobust CRISPR / Cas9 system for convenient high-efficiency multiplex genome editing in monocot and diocot plants. Mol. Plant.8, 1274-1284."), the intermediate vector pYLsgRNA- OsU6aL (disclosed in the literature "Ma et al.2015, A robust CRISPR / Cas9 system for convenient high-efficiency multiplex genome editing in monocot and diocotplants.Mol.Plant.8,1274-1284."), pYLsgRNA-OsU3aL (already In the literature "Ma et al.2015...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com