Detection method for O antigen molecular typing of citrobacter O4 and O12 serotype
A technology of citric acid bacteria and detection method, applied in the direction of microorganism-based method, biochemical equipment and method, microorganism measurement/inspection, etc., to achieve the effect of simple operation, strong repeatability and high accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Primer design
[0028] 1. Screening of specific genes
[0029] The O-antigen processing genes wzy, wzx, wzm, and wzt are highly serotype-determining and thus have been widely used as target genes for molecular serotyping of many Gram-negative bacteria. Citrobacter O12, select wxya as a specific gene. But in Citrobacter O4, because there are no specific genes like wzx / wzy, only relatively specific genes can be selected, usually some rare monosaccharide genes or glycosyltransferase genes, O4 serotype is the selection Glycosyltransferase gene GT was used as a specific gene.
[0030] In the present invention, the all_vs_all_blast method is performed on all the genes in the gene cluster for comparison, and the matching number of the specific gene must be much smaller than that of the conservative gene. Combine the above methods to find specific genes and design primers for them.
[0031] 2. Design of primers
[0032]The primers for LAMP were designed using the selecte...
Embodiment 2
[0038] Extraction of sample nucleic acid (crude extract of a pure culture of a sample isolated from any environment suitable for the life of Citrobacter)
[0039] 1. Dilute the sample to be tested with sterile water, usually the dilution factor is 1:10 (for example, 10g solid sample or 10ml liquid sample is dissolved in 90ml sterile water) to make a bacterial liquid mother solution (only the liquid is taken).
[0040] Dilute the stock solution with sterile water for another 5 gradients: 1 × 10 ‐1 , 1×10 ‐2 , 1×10 ‐3 , 1×10 ‐4 , 1×10 ‐5 , each gradient bacterial solution was evenly spread on LB solid medium and cultured at 37°C. When a single colony grows on the plate, pick up the colony and inoculate it in LB liquid medium, and cultivate overnight at 37°C and 180rpm (this step of inoculation is performed in a super-clean bench).
[0041] 2. Sample processing: Take 1 mL of overnight cultured bacterial liquid and add it to a 1.5 mL centrifuge tube, centrifuge at 8000 rpm fo...
Embodiment 3
[0052] Positive and Specific Detection
[0053] The extracted nucleic acid solution is used as the template for the LAMP reaction, the LAMP reaction system and reaction conditions:
[0054] 1. Example of LAMP reaction system (25 µL):
[0055]
[0056] 2. LAMP reaction conditions:
[0057]
[0058] 3. Test results:
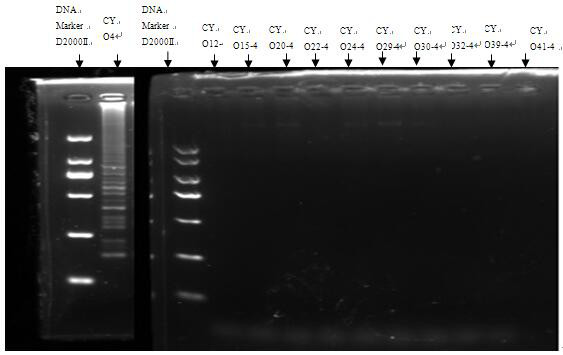
[0059] See figure 1 As shown, the genomes of Citrobacter O12, O15, O20, O22, O24, O29, O32, O39 and O41 were added to the LAMP system of Citrobacter genome sample O4, and the LAMP primers of Citrobacter O4 had good specificity.
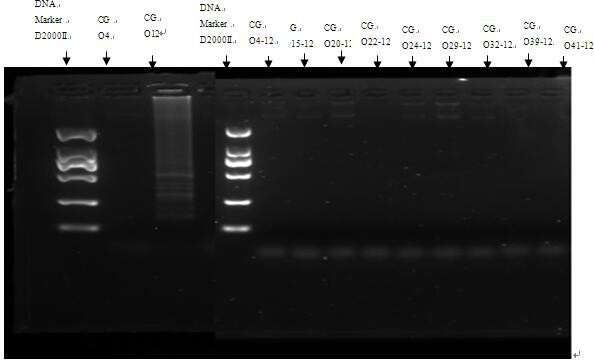
[0060] See figure 2 As shown, the genomes of Citrobacter O4, O15, O20, O22, O24, O29, O32, O39 and O41 were added to the LAMP system of Citrobacter genome sample O12, and the LAMP primers of Citrobacter O4 had good specificity.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com