Method for constructing TAP gene-deleted pig T2 cell by using CRISPR/Cas9 system
A tap2-sgRNA-2, tap1-sgRNA-2 technology, applied in urinary tract/kidney cells, genetically modified cells, cells modified by the introduction of foreign genetic material, etc. The establishment of porcine-derived T2 cell lines and epitope screening are time-consuming and labor-intensive, achieving the effect of easy transformation, convenient operation and less workload
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
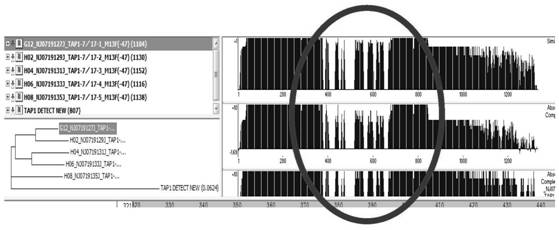
[0073] Example 1 Recovery and Karyotype Detection of PK15 Cells
[0074] Take out the cryopreservation tube from the liquid nitrogen and immediately place it in a 37°C water bath for shaking to melt the frozen cells rapidly. Transfer the thawed frozen cells into a 15mL centrifuge tube with 9mL of fresh medium, centrifuge at 1500rpm for 3min, discard the supernatant, add preheated fresh DMEM culture medium and resuspend. Add the resuspended PK15 cells to a 25 cm medium containing 4 mL of DMEM complete medium 2 In the cell culture flask, shake gently to mix, in CO 2 cultured in an incubator. After 24 hours of cultivation, it can grow to 25cm 2 Cell culture flasks for subsequent experiments. Cells were typed using spectral karyotyping techniques:
[0075] Karyotype analysis:
[0076] Freshly cultured cells were treated with colchicine to obtain more metaphases, and then treated with hypotonicity and fixation. After the specimens were made, they were stained with Giemsa sta...
Embodiment 2
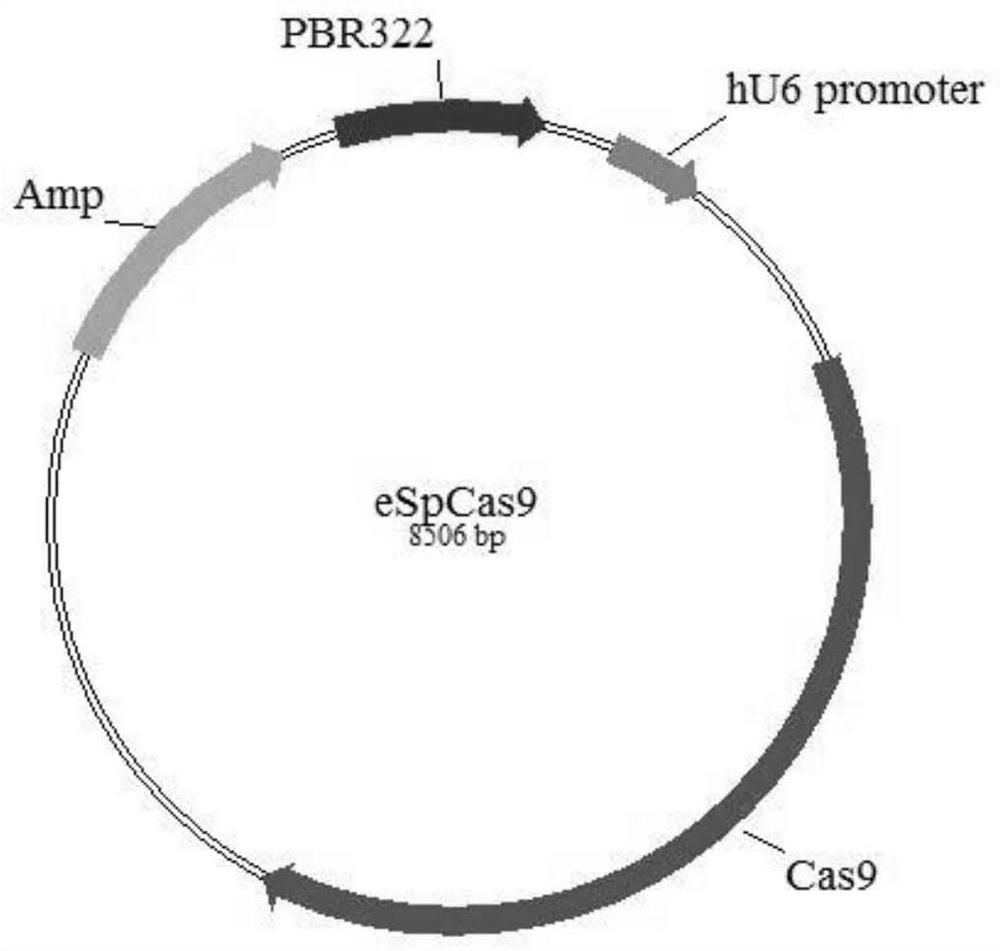
[0084] Example 2 Construction of TAP2 Knockout PK15 Cell Line Using CRISPR / Cas9 Technology
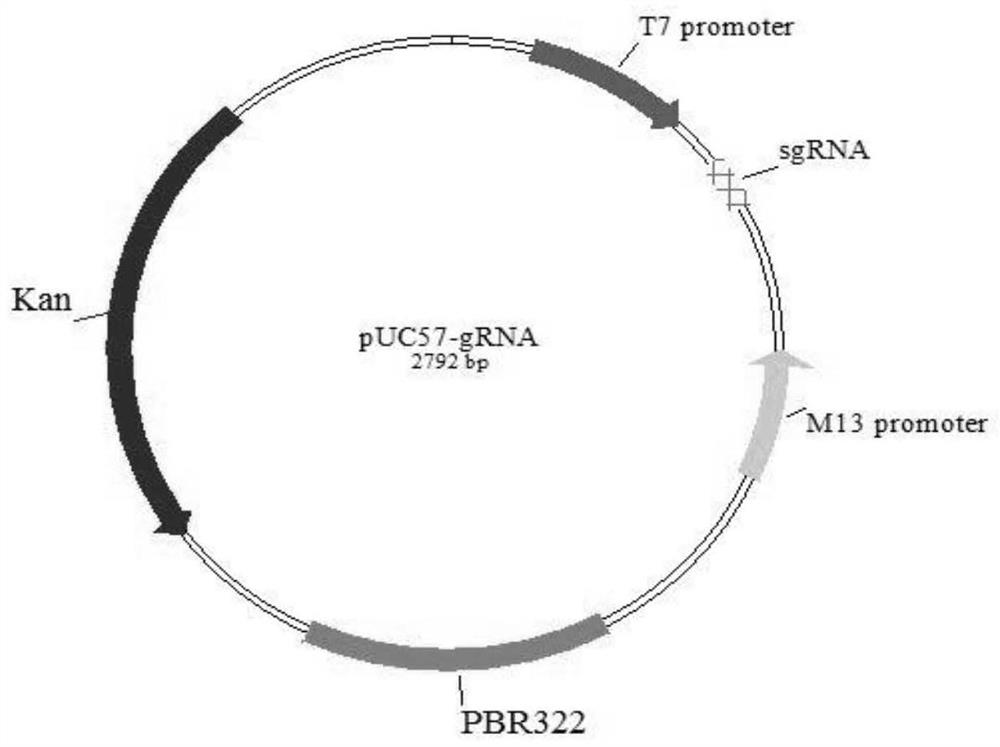
[0085] 1 Construction of pUC57-T7-sgRNA vector
[0086] (1) Design and synthesis of TAP2 gene target site
[0087] The TAP2 gene information was obtained through the Ensembl genome database in NCBI, and it has been found that there are two transcripts of this gene, both of which encode proteins. Log on to the GenBank website, download the TAP2 gene sequence of pig PK15 cells and obtain the second exon sequence (Gene ID: 733650), and design TAP2 according to the target prediction website CCtop (http: / / crispr.cos.uni-heidelberg.de) 3 pairs of sgRNA sequences of the second exon of the gene. CACC was added to the 5' end of the upstream primers of the two pairs of oligonucleotide sequences, and AAAC was added to the 5' end of the downstream primers. The TAP2-sgRNA design is shown in Table 1.
[0088] Table 1 sgRNA sequence
[0089]
[0090] Send 3 pairs of designed sgRNA sequences to...
Embodiment 3
[0141] Example 3 Construction of TAP1 Knockout PK15 Cell Line Using CRISPR / Cas9 Technology
[0142] 1 PUC57-sgRNA vector construction
[0143] (1) Design and synthesis of TAP1 gene target site
[0144] The porcine TAP1 gene transcript information was obtained through the Ensembl database in NCBI, TAP1ENSSSCG00000025618, and there are two transcripts of the TAP1 gene, namely TAP1-201 and TAP1-202. Using the second exon of the TAP1 gene as the knockout sequence, 4 pairs of oligonucleotide target recognition sequences were designed using the sgRNA prediction website (http: / / crispr.cos.uni-heidelberg.de). Add CACC to the 5' end of the upstream primer of the four pairs of oligonucleotide sequences, and add AAAC to the 5' end of the downstream primer. See Table 2 for the 4 pairs of sgRNA sequences.
[0145] Table 2 sgRNA sequence
[0146]
[0147] Send 4 pairs of designed sgRNA sequences to the company for synthesis, and anneal the synthesized sgRNA sequences to form double s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com