Detection probe, preparation method thereof, and application
A technology for detecting probes and sequencing, which is used in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., which can solve the performance degradation of probe coverage uniformity, difficult preservation of RNA probes, and capture reactions. Cost increase and other issues, to achieve the effect of reducing experimental cost, high uniformity and low cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
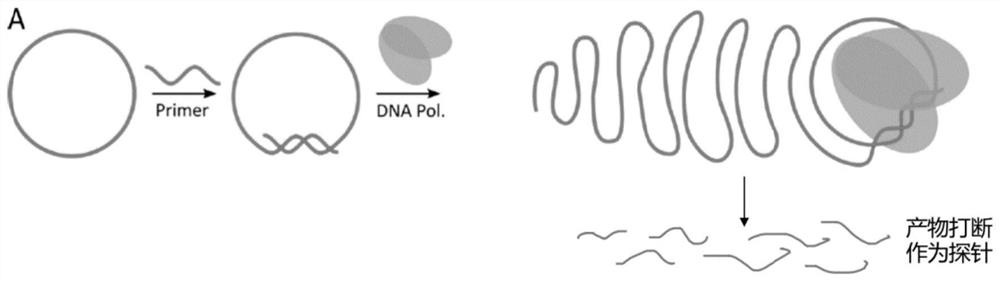
[0041] A kind of embodiment of DNA probe prepared by the method of the present invention (the schematic diagram of the preparation principle of DNA probe is as follows figure 1 ), the preparation method is as follows:
[0042] 1. Preparation of Minicircle plasmid containing the target fragment:
[0043] (1), using primer 1: ccatgtaccaatgttgcagt (SEQ ID NO.3) and primer 2: ggatcctttgccccagtgtt (SEQ ID NO.4), by PCR method, from pBR322-HPV16 (purchased from ATCC, trade name: human papilloma virus type 16 45113 TM ) was cloned to obtain the full-length fragment of HPV16; the PCR system is shown in Table 1. KAPA HiFi Hot Start Ready Mix was purchased from Roche.
[0044] Table 1 HPV16 full-length fragment amplification PCR system
[0045] components Dosage pBR322-HPV16 plasmid 1μL Primer 1 (10 μM) 1μL Primer 2 (10 μM) 1μL KAPA HiFi Hot Start Ready Mix(2×) 25 μL water 22 μL total capacity 50μL
[0046] The reaction pr...
Embodiment 2
[0074] Embodiment 2 The method for the probe of the present invention to carry out liquid phase capture
[0075] 1. Sample preparation stage
[0076] genomic DNA fragmentation
[0077] 1) Transfer the genomic DNA to a 0.6mL MaxyClear Snap lock Microcentrifuge Tube according to the above system, mix well, centrifuge briefly and put it on ice for use;
[0078] 2) Turn on the ultrasonic interrupter Bioruptor Pico in advance. After the temperature of the cold cycler drops to 4°C, set the parameters ON30s, OFF30s as a cycle, and every 10cycles is a round, and a total of 3 rounds are performed. After each round, the sample is placed Mix well on a shaker, and then perform the next round of fragmentation after brief centrifugation (the more thoroughly the mixing, the more concentrated the size of the fragmented sample); after fragmentation, the main peak of the sample detection is about 150bp-200bp.
[0079] 2. Library Construction Phase
[0080] 1. End repair, add "A" to the 3' en...
Embodiment 3
[0144] An embodiment of RNA probe prepared by the method of the present invention (the schematic diagram of the preparation principle of RNA probe is as follows figure 1 ), except for rolling circle amplification and purification, the rest of the process is the same as in Example 1 (note: the product after RCT is RNA, and all consumables used in the experimental process, including pipette tips, centrifuge tubes, interrupted tubes, etc., need to be kept No RNase. Change gloves frequently during operation to avoid RNase contamination.).
[0145] Rolling circle amplification and purification:
[0146] Prepare the reaction system according to the table below (the relevant reagents are purchased from NEB, product number T7 RNA polymerase).
[0147] Table 11 Reaction system
[0148] components Dosage 10×T7 RNA polymerase buffer 2μL RCA primer 2.5μL Sample to be amplified 5ng Enzyme-free water Rehydrate to 14.2 μL
[0149] Incubate at 95...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com