Method for preparing cyclic adenosine monophosphate by adenylate cyclase
A technology of adenylate cyclase and cyclic adenosine monophosphate, applied in biochemical equipment and methods, botanical equipment and methods, enzymes, etc., can solve the problem of high-efficiency expression mechanism that has not been clarified, further research is needed, and the yield is low and other problems, to achieve the effect of short cycle, mild conditions and simple industry
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
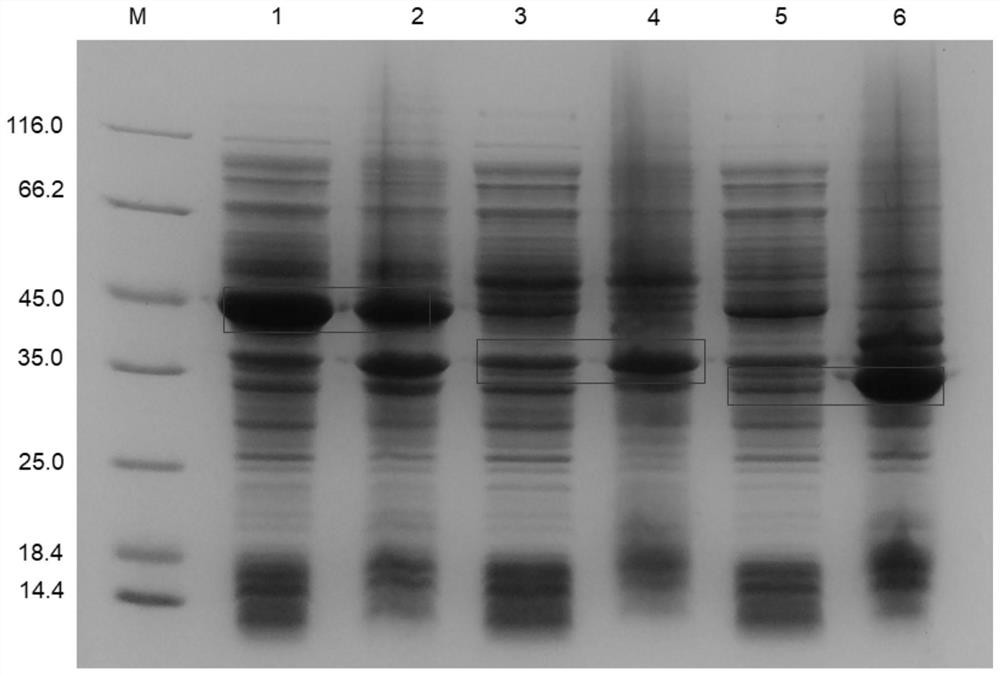
[0056] Screening for efficient adenylyl cyclase (AC)
[0057] Using the amino acid sequence of known adenylyl cyclase (asAC) as a template, six potential adenylyl cyclase sequences were obtained by NCBI-Blastp online alignment, namely ag30AC, gmc3AC, naAC, kiAC, biAC and tcAC , the homology between the screened six potential adenylyl cyclases and the template adenylyl cyclase was 89%, 77%, 65%, 56%, 46%, 36%, respectively. The amino acid sequences and nucleotide sequences of the six screened adenylyl cyclases are shown in Table 1.
[0058] Table 1
[0059]
[0060]
[0061] Subsequently, the corresponding nucleotide sequences were synthesized in vitro and connected into the expression vector pET28a to obtain six recombinant plasmids pET28a-ag30AC, pET28a-gmc3AC, pET28a-naAC, pET28a-kiAC, pET28a-biAC and pET28a-tcAC, which will be obtained 6 kinds of recombinant plasmids were transformed into E.coli BL21(DE3), spread on LB plates containing 50 μg / mL kanamycin, and scree...
Embodiment 2
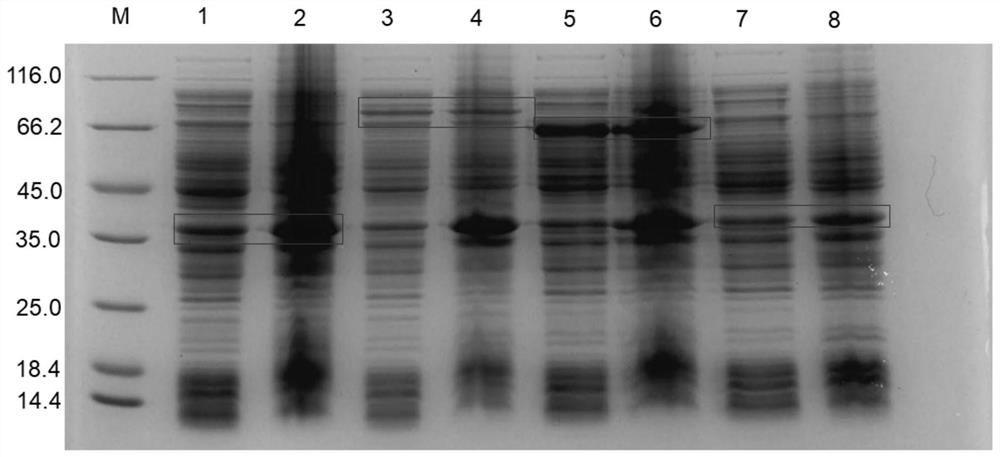
[0067] IPTG induced expression of adenylate cyclase
[0068] The recombinant bacteria E.coli BL21(DE3) / pET28a-ag30AC, E.coli BL21(DE3) / pET28a-gmc3AC and E.coli BL21(DE3) / pET28a-tcAC screened in Example 1 were selected to inoculate to 50 μg / In the LB medium of mL kanamycin, culture at 37°C and 220rpm for 8 hours, inoculate into fresh LB medium at 2% inoculum size, and cultivate to OD at 37°C and 220rpm 600 When it reaches about 0.6, add IPTG with a final concentration of 0.5mM, induce expression at 28°C and 220rpm for 14h, then centrifuge at 4°C and 8000rpm / min for 10 minutes to obtain wet cells containing adenylyl cyclase. Subsequently, the wet bacteria were resuspended with pure water, and crushed according to the crushing method provided in Example 1 to obtain a crude enzyme solution to measure the enzyme activity.
[0069] The enzyme activity assay system was as described in Example 1. After testing, the enzyme activities of ag30AC, gmc3A, and tcAC induced by IPTG were 14...
Embodiment 3
[0071] Optimization of Catalytic Conditions for Adenylyl Cyclase
[0072] According to the method described in Example 2, the expression of recombinant bacteria E.coli BL21(DE3) / pET28a-tcAC was induced, and the crude enzyme solution was obtained after centrifugation and crushing, and the optimum temperature in the catalytic process was optimized.
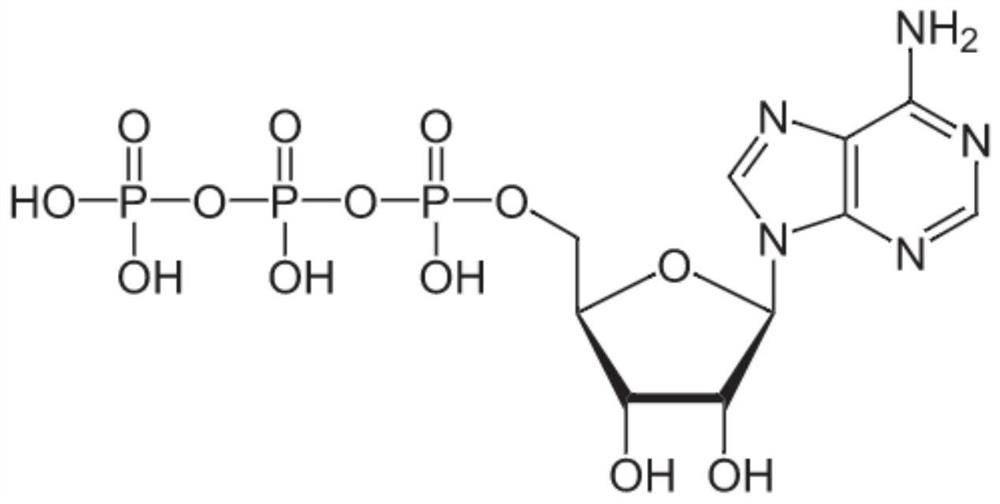
[0073] The optimization system was carried out in a 1mL reaction tube. Take 900 μL of the ATP substrate solution prepared in Example 1 as the base solution, incubate it with the crude enzyme solution at 25-90°C for 5 minutes, and then add 100 μL of the crude enzyme solution (based on the volume before crushing). Wet bacterial concentration meter, the final concentration is 10g / L), react in a water bath at 25-90°C for 15min, take 100μL of the reaction solution and add 5μL of 6M hydrochloric acid solution to terminate the reaction. Add 900 μL of pure water to dilute 10 times, mix well, then take 100 μL of it and add 900 μL of pure wat...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com