Compositions and methods for increasing ethanol production by yeast using gcy1 and dak1
A technology of yeast and Saccharomyces, which is applied in the field of glycerol dehydrogenase-dihydroxyacetone kinase bifunctional fusion polypeptide, which can solve the problems affecting the production rate of ethanol and disadvantages
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0181] Materials and methods
[0182] Liquefaction preparation
[0183] The liquefied product (ground corn slurry) was added by adding 600ppm urea, 0.124SAPU / g ds FERMGEN TM Prepared with 2.5X (acid fungal protease), 0.33 GAU / g ds TrGA variant glucoamylase, and 1.46 SSCU / g ds GC626 (Aspergillus alpha-amylase), adjusted to pH 4.8.
[0184] AnKom assay
[0185] 300 μL of the concentrated overnight yeast culture was added to each of the multiple ANKOM bottles filled with 30 g of the prepared liquefaction to achieve a final OD of 0.3. The flasks were then incubated for 65 hours at 32°C with shaking (150 RPM).
[0186] HPLC analysis
[0187] Samples from serum bottles and AnKom experiments were collected in Eppendorf tubes by centrifugation at 14,000 RPM for 12 minutes. The supernatant was filtered with a 0.2 μΜ PTFE filter and then used for HPLC (Agilent Technologies 1200 series) analysis under the following conditions: Bio-Rad Aminex HPX-87H column operating at 55°C. 0.6ml...
example 2
[0191] Construction of a plasmid carrying a fusion gene encoding N-terminal glycerol dehydrogenase and C-terminal dihydroxyacetone kinase
[0192] Synthetic GCY1-L1-DAK1 and GCY1-L2-DAK1 are fusion genes comprising codon-optimized glycerol fused via linker 1 (SEQ ID NO:3) and linker 2 (SEQ ID NO:4), respectively Dehydrogenase (GCY1, SEQ ID NO:2) and dihydroxyacetone kinase (DAK1, SEQ ID NO:1). The amino acid sequences of fusion polypeptides GCY-L1-DAK1 and GCY1-L2-DAK1 are represented by SEQ ID NO: 5 and 6, respectively.
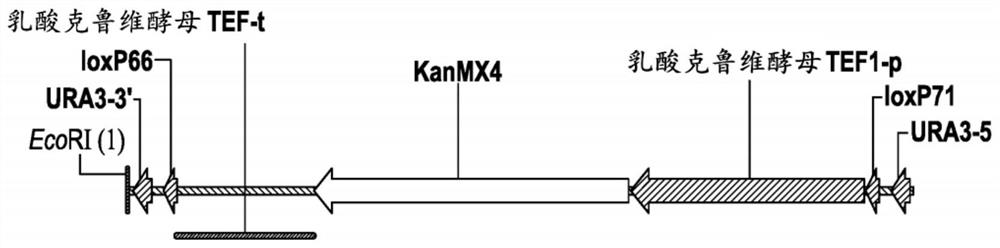
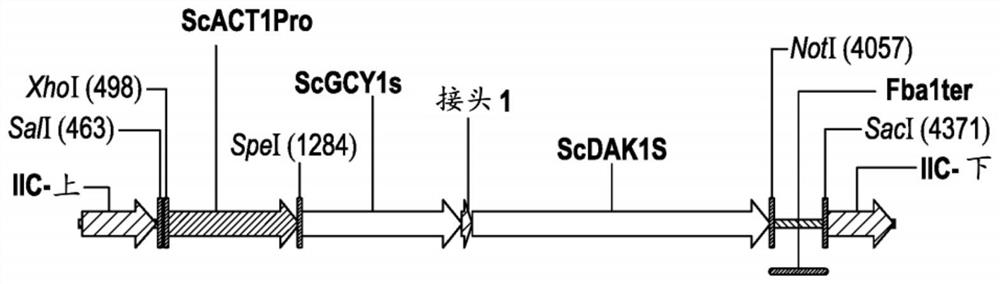
[0193] Plasmid pZKIIC-YL1K contains an expression cassette to express the GCY1-L1-DAK1 fusion polypeptide under the control of the ACT1 promoter (YFL039C locus) and the FBA1 terminator (YKL060C locus). Plasmid pZKIIC-YL1K was designed to integrate the expression cassette into Saccharomyces chromosome II at positions 345856 and 350891. The functional and structural composition of plasmid pZKIIC-YL1K is described in Table 2.
[0194] Table 2. Functional and...
example 3
[0198] Construction of a plasmid with a fusion gene encoding N-terminal dihydroxyacetone kinase and C-terminal glycerol dehydrogenase
[0199] Synthetic DAK1-L1-GCY1 and DAK1-L2-GCY1 are fusion genes comprising codon-optimized two genes fused by linker 1 (SEQ ID NO:3) and linker 2 (SEQ ID NO:4), respectively. Hydroxyacetone kinase (DAK1, SEQ ID NO:1) and glycerol dehydrogenase (GCY1, SEQ ID NO:2). The amino acid sequences of the fusion polypeptides DAK-L1-GCY1 and DAK1-L2-GCY1 are represented by SEQ ID NO: 7 and 8, respectively.
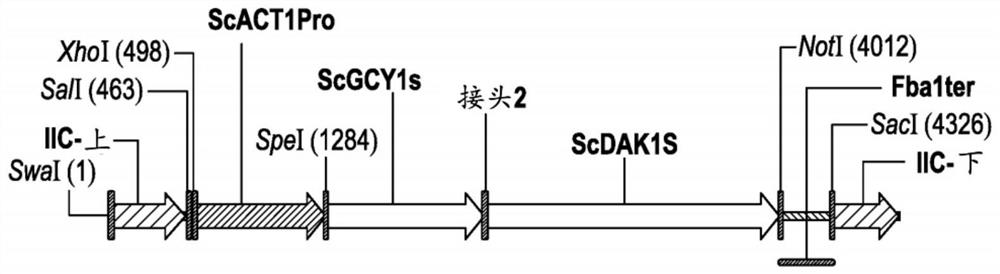
[0200] Plasmids pZKIIC-KL1Y and pZKIIC-KL2Y contain expression cassettes to express DAK-L1-GCY1 and DAK1-L2-GCY1 fusion polypeptides under the control of the ACT1 promoter (YFL039C locus) and the FBA1 terminator (YKL060C locus). Both pZKIIC-KL1Y and pZKIIC-KL2Y were designed to integrate the expression cassette into Saccharomyces chromosome II at positions 345856 and 350891. The functional and structural composition of plasmids pZKIIC-KL1Y and pZKI...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com