Multiple nucleic acid detection method, combination and kit
A multiple nucleic acid and signal detection technology, applied in the field of molecular biology, can solve the problems of increased detection cost and unfavorable clinical promotion and use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0177] Example 1 is designed to detect primer sets and flexible probes for respiratory-associated pathogens
[0178] In order to verify the combination of the present invention in the detection of influenza A virus (Influenza A virus, IAV), influenza B virus (Influenza B virus, IBV), respiratory syncytial virus and (Respiratory Syncytial Virus, RSV), adenovirus (Adenovirus, ADV), Mycoplasma pneumoniae (Mycoplasma pneumonia, MP) and novel coronavirus (SARS-CoV-2), designed the primers and probes in Table 1 below.
[0179] Table 1
[0180]
[0181] Wherein, upstream primer 1 and downstream primer 1 are specific primers designed for the target sequence of adenovirus; upstream primer 2 and downstream primer 2 are specific primers designed for the target sequence of influenza B virus; upstream primer 3 and downstream primer 3 are Specific primers designed for the target sequence of Mycoplasma pneumoniae; upstream primer 4 and downstream primer 4 are specific primers designed fo...
Embodiment 2
[0185] Example 2 Single detection of adenovirus, influenza B virus, respiratory syncytial virus, influenza A virus, Mycoplasma pneumoniae and novel coronavirus
[0186] Sample preparation: Use the in vitro transcribed RNA of each target sequence as a positive sample, and test each target sequence separately to verify the detection ability of a single virus infection. Pure water was used as a no-template control (NTC).
[0187] Reaction preparation:
[0188] After the sample preparation is completed, configure the reaction system according to the proportions described in Table 3 below.
[0189] table 3
[0190]
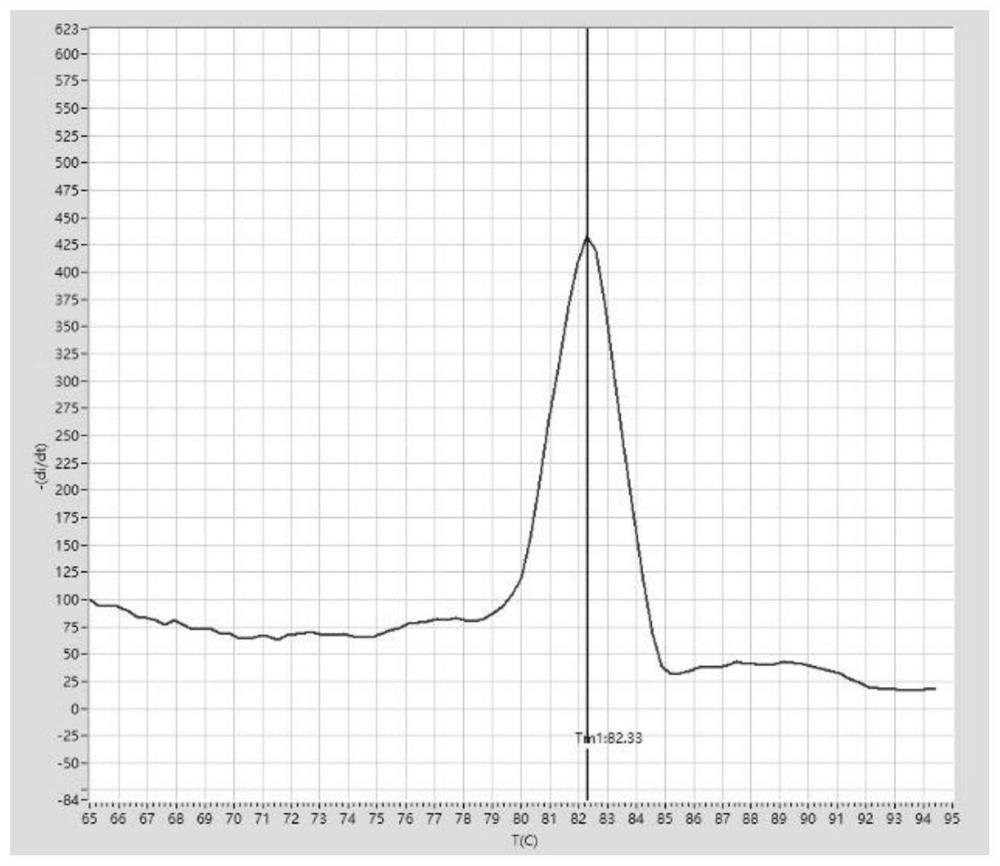
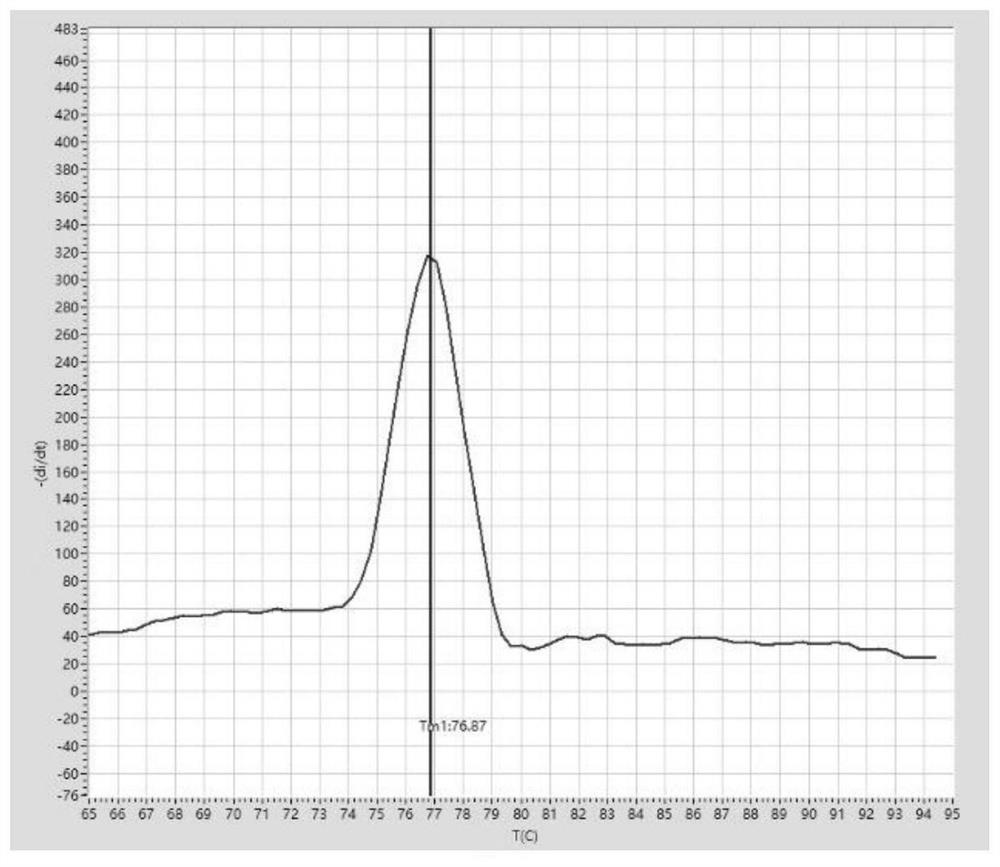
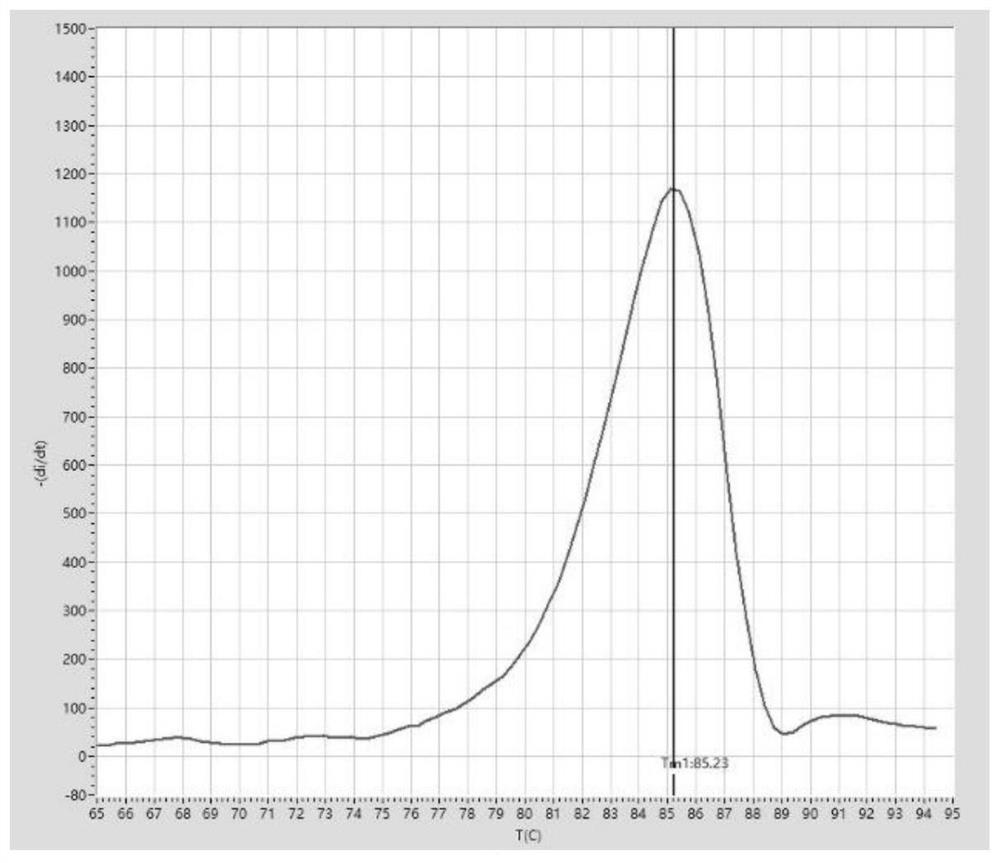
[0191] PCR amplification and melting curve analysis:
[0192] After sealing the PCR tube cap, mix the sample gently, then centrifuge briefly and let it stand at room temperature for 5 minutes. The PCR tube was placed in the palm centrifuge again, and after a short centrifugation, it was transferred to the tray of a fluorescent quantitative PCR instrument (Suzhou...
Embodiment 3
[0197] Example 3 Multiple detection of adenovirus, influenza B virus, respiratory syncytial virus, influenza A virus, Mycoplasma pneumoniae and novel coronavirus
[0198] Sample preparation: Use the in vitro transcribed RNA of each target sequence as a positive sample, mix the two target sequence templates of the same fluorescent channel in Table 2, and then perform detection to verify the detection ability of virus mixed infection. Ultrapure water was used as a no-template control (NTC).
[0199] Reaction preparation:
[0200] After the sample preparation is completed, configure the reaction system according to the proportions described in Table 3.
[0201] PCR amplification and melting curve analysis:
[0202] After sealing the PCR tube cap, mix the sample gently, then centrifuge briefly and let it stand at room temperature for 5 minutes. The PCR tube was placed in the palm centrifuge again, and after a short centrifugation, it was transferred to the tray of a fluorescent...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com