Method for improving hydrolase robustness in combination with high-pressure molecular dynamics simulation and free energy calculation
A molecular dynamics, hydrolytic enzyme technique, applied in the field of computational chemistry
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Example 1: T1 lipase
[0054] 1 Screening of highly robust related regions and sites
[0055] In this example, the crystal structure (PDB id: 2dsn) of wild-type T1 lipase (T1 lipase from Geobacillus zalihae strain T1, T1 lipase) was used as the initial model, and the mutant was modeled using SWISS-MODEL. Gromacs (version 2019.03) performs molecular dynamics simulations.
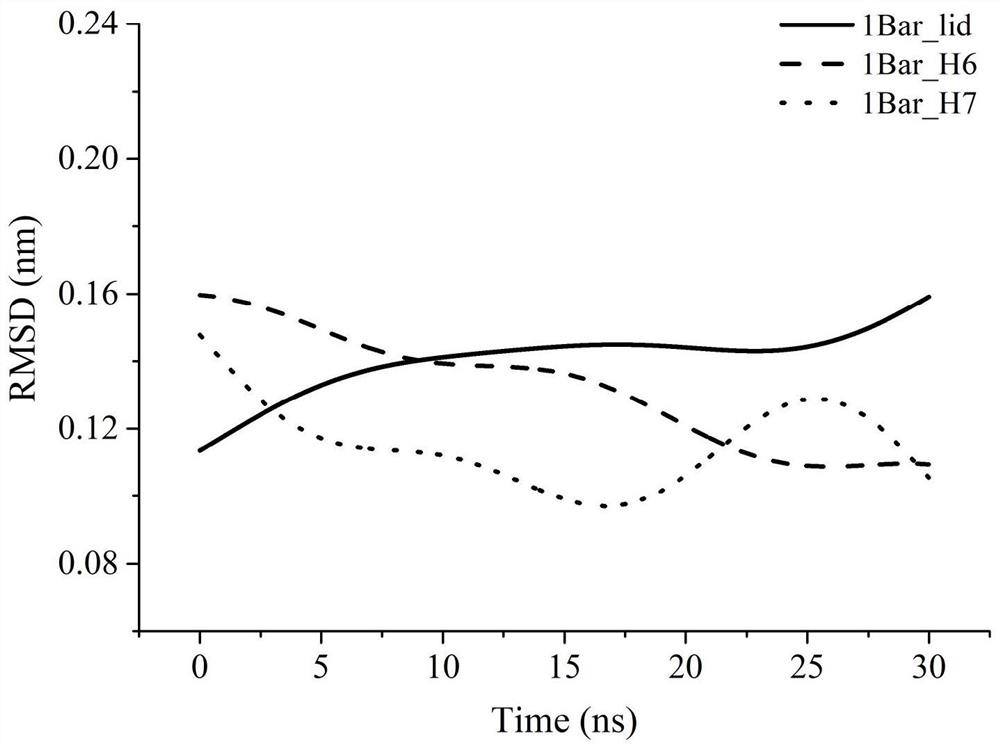
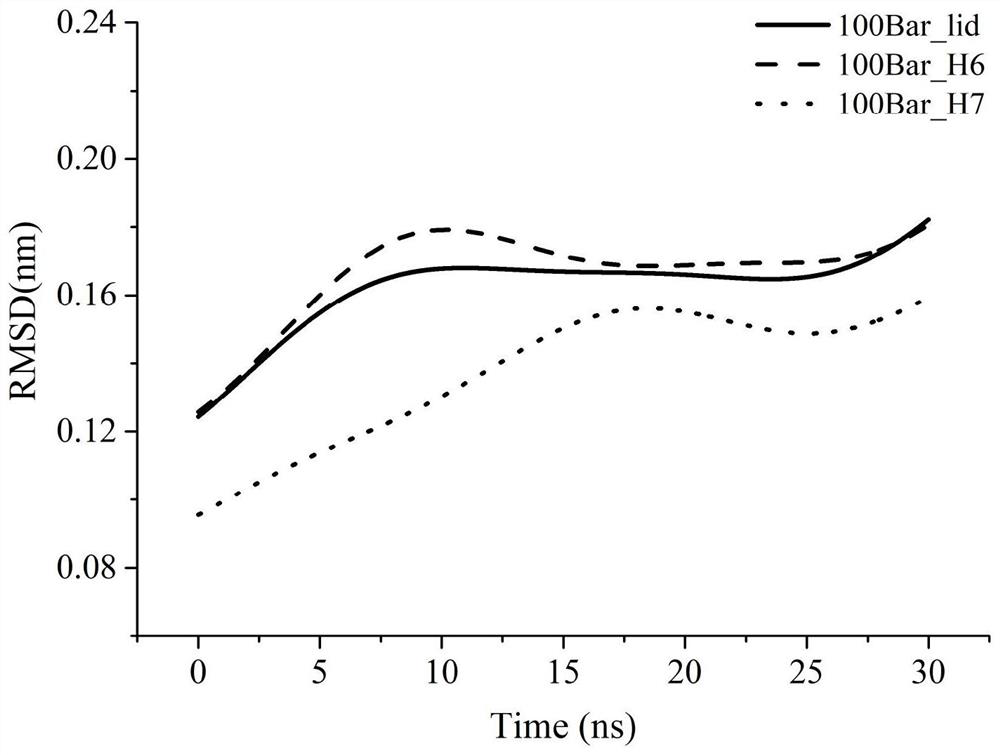
[0056] The shortest distance from the edge of the box is 1.0nm, and the water model uses TIP4P, because this model is one of the most reliable water models for studying pressure effects, and then 5 Na + Neutralizing the charge brings the entire system into equilibrium. The 50000-step steepest descent method is used to minimize the energy of the system to ensure that the structure is normal, the distance between atoms is appropriate, and the geometric configuration is reasonable, and then a 400ps constant temperature and constant volume ensemble (NVT) balance is performed under periodic boundary conditi...
Embodiment 2
[0101] Embodiment 2: Rhizomucor miehei lipase (RML lipase)
[0102] 1 Screening of highly robust related regions and sites
[0103] The crystal structure of RML lipase (PDB id: 4tgl) was downloaded from the Protein Data Bank as the research object, and the downloaded crystal structure was corrected and optimized with the Repair and Optimize programs in FoldX suite 5.0. The MD simulation uses the GROMACS 2019.03 installation package. The AMBER99 force field is applied during the simulation. The protein is placed in a cubic box filled with water, and the distance between the protein and the edge of the box is at least 1.0nm. The water model uses TIP4P. Since this model is a research One of the most reliable water models for pressure effects, then add 10 sodium ions for charge balance. The steepest descent method with 50,000 steps is used to minimize the system to ensure normal structure, proper distance between atoms, and reasonable geometric configuration. Then, under periodic...
Embodiment 3
[0116] Example 3: Carbamate Hydrolase
[0117] 1 Screening of highly robust related regions and sites
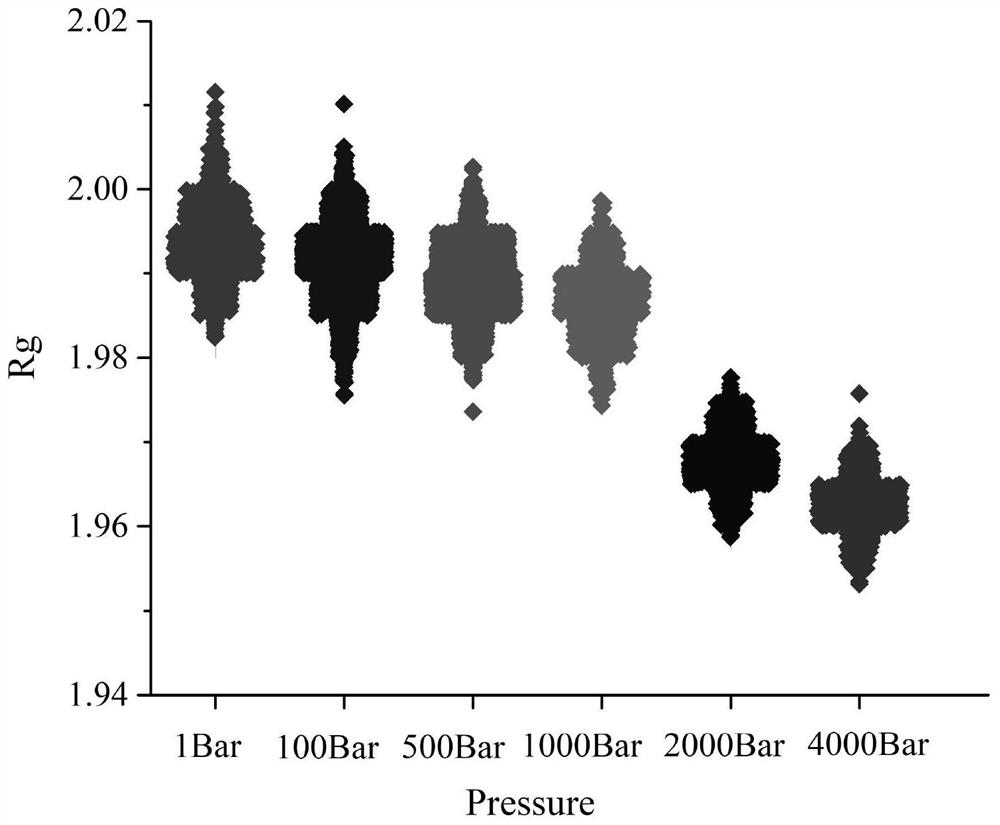
[0118] The templates with high homology in the PDB database were used to model separately, and the Glutamyl-tRNA(Gln)amidotransferase subunit A (PDBID :2G5H) as a template, use the online three-dimensional structure simulation program SWISS-MODEL (http: / / swissmodel.expasy.org / interactive) to urethane hydrolase (EC 3.5.1.75, Urethanase, its amino acid sequence is shown in the sequence listing, Derived from Lysinibacillus fusiformis SC02) for three-dimensional structure modeling, under the conditions of 1Bar, 100Bar, 500Bar, 1000Bar, 2000Bar, 4000Bar pressure and 313K, molecular dynamics simulations were performed on UH, and the search for enzyme molecules Instability-associated structures or regions. Analysis of the B-factor value of each amino acid of the UH protein by FlexServ software shows that the B-factor response value of Leu327 in the sequence is the highest, indica...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com