Construction method and application of saccharomyces cerevisiae capable of dynamically regulating 7-deoxycholesterol and vitamin D3
A technology of Saccharomyces cerevisiae and construction method, applied in the direction of microorganism-based methods, biochemical equipment and methods, applications, etc., can solve the problems of harsh reaction conditions, unfavorable sustainable development, high energy consumption, etc., and achieve the effect of inhibiting branch paths
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
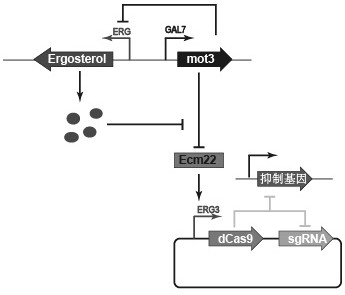
[0038] Example 1 Construction of the first Saccharomyces cerevisiae DC-1
[0039] (1a) Using the S228C genome of Saccharomyces cerevisiae as a template, the gene fragment Mot3, the gene fragment GAL7, and the upstream homology arm gene fragment NAT were amplified by primers, and the gene fragment Loxp was obtained by using the plasmid pMHyLp-TRP1 as a template. -Trp.
[0040] The primers for the amplification of the gene fragment Mot3 are shown in SEQ ID 1 and SEQ ID 2, the primers for the amplification of the gene fragment GAL7 are shown in SEQ ID 3 and SEQ ID 4, and the amplification of the primers for the upstream homology arm gene fragment NAT The primers are shown in SEQ ID 5 and SEQ ID 6, and the primers for amplifying the gene fragment Loxp-Trp are shown in SEQ ID 7 and SEQ ID 8.
[0041] (1b) Make the original yeast competent, transform the four gene fragments obtained in step (1a) into Saccharomyces cerevisiae, and use the primers shown in SEQ ID 9 and SEQ ID 10 to p...
Embodiment 2
[0043] Example 2 Construction of a dCas9 system dynamically regulated by ergosterol
[0044] (2a) Artificially synthesized gene fragment PERG3-dCas9-Tcyc1-PSNR52-sgRNA-TADH1, using the primers shown in SEQID 11 and SEQ ID 12 to amplify the gene fragment Dcas9-sgRNA, using the plasmid PY13-TEF1 plasmid as a template, using The primers shown in SEQ ID 13 and SEQ ID 14 were amplified to obtain a partial plasmid fragment PY13-1.
[0045] (2b) The gene fragment Dcas9-sgRNA obtained in step (2a) and the gene fragment PY13-1 were seamlessly connected using seamless cloning enzyme. After reacting in a water bath at 50°C for 30 minutes, take 10 μl of the reaction sample and add it to 50-100 μl JM109 Escherichia coli competent, mix gently, and incubate on ice for 30 minutes. Heat shock in a water bath at 42°C for 90 seconds, quickly put it in an ice bath, let it stand for 2-3 minutes, add 500μl LB medium, incubate at 37°C for 1 hour, centrifuge at 5000rpm for 1 minute, remove 500μl med...
Embodiment 3
[0047] Example 3 Construction of the second Saccharomyces cerevisiae DC-2
[0048] (3a) Artificially synthesized gene fragment P TEF1 -DHCR24-T cyc1 -P GAP -DIC-T ADH1 , using the S228C genome of Saccharomyces cerevisiae as a template, using the primers shown in SEQ ID 17 and SEQ ID 18 to amplify to obtain the upstream homology arm gene fragment gal80F, using the primers shown in SEQ ID 19 and SEQ ID 20 to amplify The downstream homology arm gene fragment gal80R was obtained, and the pMHyLp-leu plasmid was used as a template to amplify the gene fragment loxp-Leu using the primers shown in SEQ ID 21 and SEQ ID 22.
[0049] (3b) The four fragments P obtained in step (3a) TEF1 -DHCR24-T cyc1 -P GAP -DIC-T ADH1 , gal80F, gal80R and loxp-Leu gene fragments were overlap-extended PCR, and after 1% agarose gel electrophoresis was verified to be correct, the fragments were recovered by cutting the gel to obtain the fusion gene fragment gal80F-loxT-P TEF1 -DHCR24-T cyc1 -P GAP...
PUM
| Property | Measurement | Unit |
|---|---|---|
| wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com