Method for preparing recombinant serratia marcescens nuclease
The technology of Serratia nuclease and Serratia nucleic acid is applied in the field of biomedicine and can solve the problems of damage to the genome of Escherichia coli, long production time and high production cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
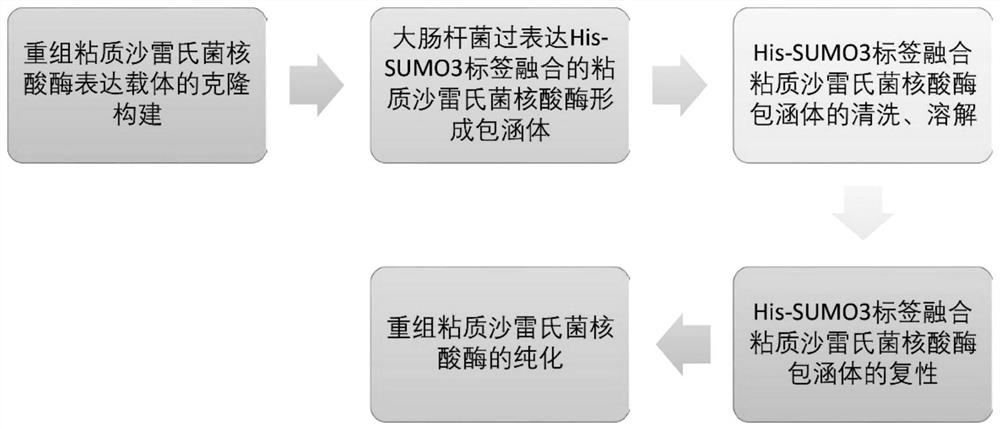
[0066] The invention provides a method for preparing recombinant Serratia marcescens nuclease with high yield, high purity, easy operation, short production cycle and low cost, which is suitable for large-scale industrial production. As a preferred embodiment, the method of the present invention includes the following steps: cloning construction of recombinant Serratia marcescens nuclease expression vector: total gene synthesis C47S mutation transformed SUMO3 tag fusion Serratia marcescens nuclease base The base fragment was introduced into the pET28a vector; the overexpressed C47S mutation engineered SUMO3 tag was fused to Serratia marcescens nuclease to form inclusion body protein.
[0067] Further, it also includes cleaning and dissolving the C47S mutation modified SUMO3 tag fusion Serratia marcescens nuclease inclusion body protein. Preferably, the cleaning solution components include: 10-50mM Tris (Tris) pH7.4, 0.1-1M NaCl, 0.1-1% Triton X-100; Preferably, the solution co...
Embodiment 1
[0086] Example 1, Construction of wild-type Serratia marcescens nuclease fusion His-SUMO3 tag clone
[0087] In order to optimize the expression of Serratia marcescens nuclease, the inventors tested various expression schemes, including linking it with His tag, linking it with His-SUMO3 tag, etc., for fusion and expression.
[0088] The amino acid sequence of the SUMO3 fusion tag is as follows (SEQ ID NO: 1):
[0089]
[0090] The amino acid sequence of the wild-type Serratia marcescens nuclease is as follows (SEQ ID NO:2; excluding its signal peptide sequence at positions 1-21):
[0091]
[0092] The present inventors connected the 92nd (102nd in SEQ ID NO: 1) glycine of the SUMO3 tag protein to the 22nd aspartic acid of Serratia marcescens nuclease (without the 1-21st signal Peptide, that is, the first position of SEQ ID NO:2), and add a His tag (6XHis) at the N-terminal to obtain His-SUMO3-Nuclease, the amino acid sequence of which is as follows (SEQ ID NO:3):
[00...
Embodiment 2
[0108] Example 2, Optimization of Refolding of Recombinant Serratia marcescens Nuclease Inclusion Body
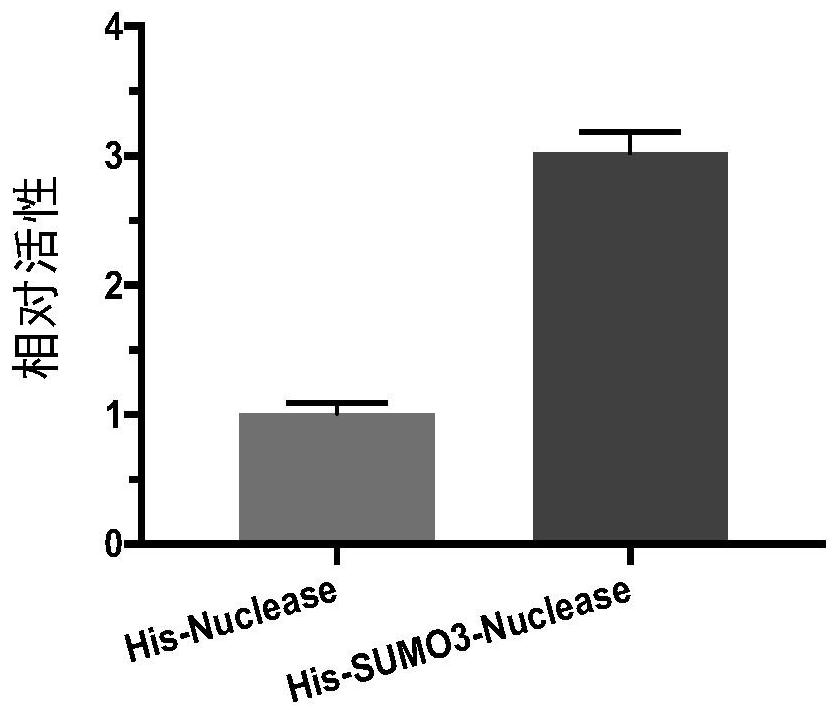
[0109] According to Example 1, the His-SUMO3 expression tag was selected for recombinant expression of Serratia marcescens nuclease. In order to further improve its renaturation rate and expression efficiency, the inventors further optimized it.
[0110] The inventors first studied the protein sequence of the Serratia marcescens nuclease, including analyzing its tertiary structure, in order to find the mutation point to improve its performance. After analysis and transformation of multiple sites and experimental verification, no sites that can effectively improve the renaturation rate of inclusion bodies were found in the experimental results.
[0111] Afterwards, the inventors attempted to modify the expression tag of His-SUMO3, and conducted research analysis and experimental verification of multiple sites in His-SUMO3. It was found that the mutation of Cys at the 47th ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com