RNA (Ribonucleic Acid) 4-mercaptouracil specific labeling method and application thereof
A mercaptouracil, RNA4- technology, applied in the field of nucleic acid chemistry, can solve the problems of low reaction efficiency, harsh reaction conditions, oxidative damage of classical bases, etc., and achieve the effect of good selectivity and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
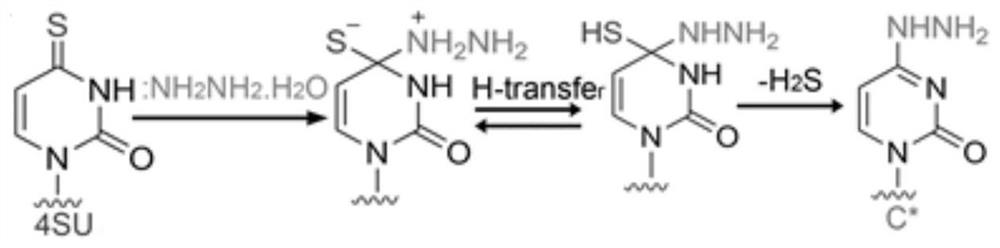
[0069] Example 1: Using the addition and elimination reaction of hydrazine hydrate to RNA 4sU according to the present invention, a structure similar to cytosine was obtained.
[0070] In vitro transcription of RNA with 4sU site, the 4 S U-RNA reacts with hydrazine hydrate, and the reaction conditions are: water as solvent, 10mM DTT buffer, heating at 50°C for 1.5h. After the reaction, the RNA was recovered by alcohol precipitation. The labeled RNA was reacted with rSAP and RNAse I at 37°C for 12 hours to degrade the nucleic acid into nucleosides. Finally, various nucleosides in the sample were detected by LC-MS / MS.
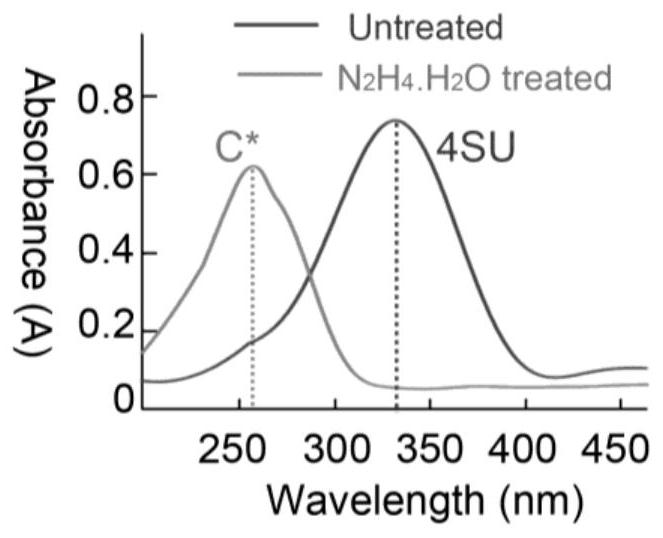
[0071] see figure 2 , for 4 S The ultraviolet absorption spectra before and after the reaction of U and hydrazine hydrate, it can be seen from this figure that hydrazine hydrate can convert 4 S U is completely reacted, and a new product cytidine analogue C* is obtained.
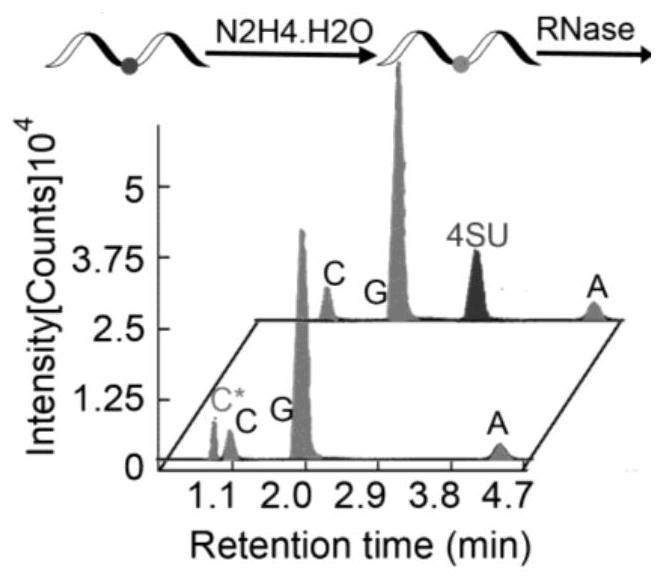
[0072] Nucleoside HPLC mass spectrometry after RNA degradation see image 3 , it can...
Embodiment 2
[0074] Example 2: Single-base recognition of 4sU using the addition and elimination reaction of hydrazine hydrate to 4sU according to the present invention
[0075] RNA with 4sU site was transcribed in vitro, and 4Su-RNA was reacted with hydrazine hydrate. The reaction conditions were water as solvent, 10mM DTTbuffer, and heating at 50°C for 1.5h. After the reaction, the RNA was recovered by alcohol precipitation. The labeled RNA was subjected to reverse transcription and PCR. PCR products were characterized by 15% PAGE and sequencing. A mutation from T to C occurred at the position of the original 4sU, therefore, this method realized the single base recognition of 4sU.
Embodiment 3
[0076] Example 3: Identification of Modification Positions of Naturally Occurring 4-Mercaptouracil
[0077] The total RNA was extracted from Escherichia coli, and the total RNA was reacted with hydrazine hydrate. The reaction conditions were water as a solvent, 10mM DTT buffer was added, and the reaction was carried out at 50°C for 1.5h. tRNA with specific primers of known sequence val Reverse transcription, PCR, and next-generation sequencing were used to identify and confirm the position of the naturally occurring modified 4-mercaptouridine.
[0078] For the process of subcloning in this example, see Figure 10 , for the results of next-generation sequencing of subcloned plaques, see Figure 11 , which shows the naturally occurring 4 S The U site is changed from the original A:T pairing to the C:T pairing, indicating that the method of the present invention can recognize naturally occurring 4 S U site.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com