Use of system in improving insertion efficiency of non-natural amino acid
A technology of unnatural amino acid and high efficiency, applied in the application field of insertion efficiency, can solve the problems of reducing inhibition and low efficiency, and achieve the effect of improving insertion efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
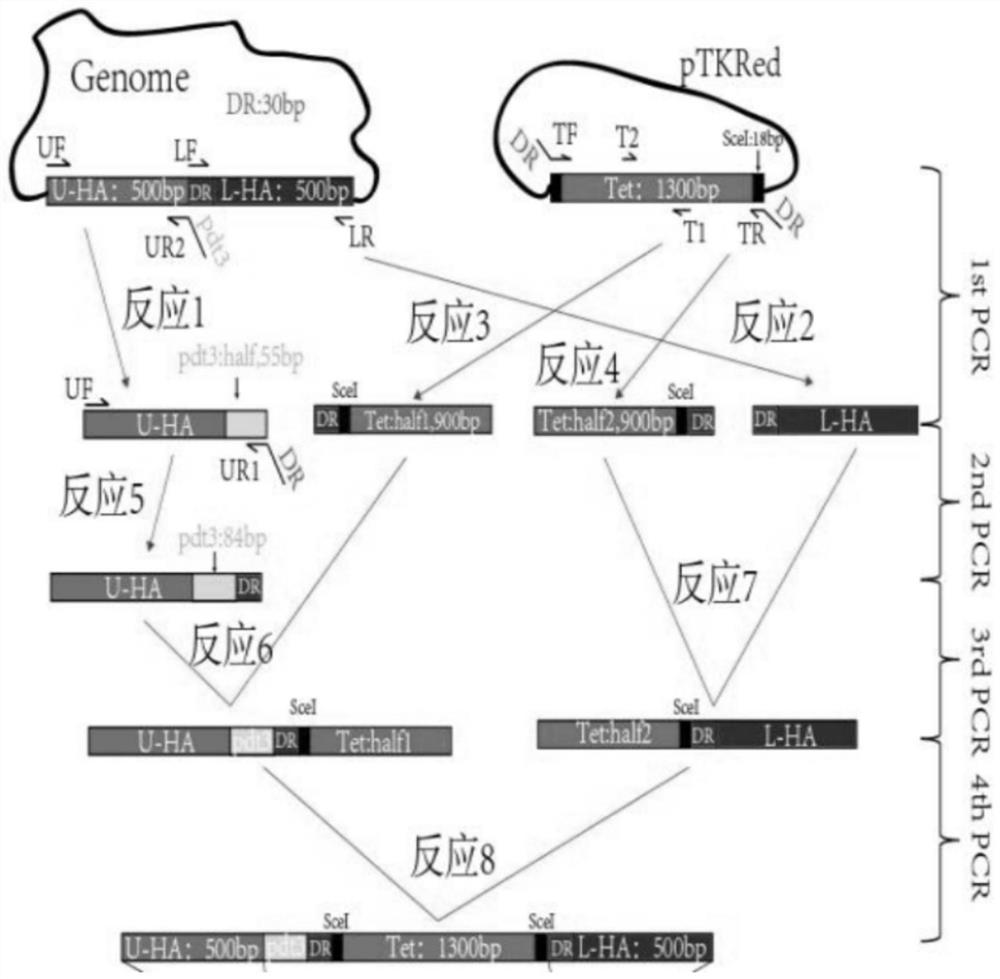
[0050] Example 1 Inserting pdt3 tags on RF1 and RF2 genes using the λ-Red system
[0051] 1. Preparation of electroporation competent:
[0052] Dilute the frozen BL21 Star (DE3) strain with LB medium at a ratio of 1:100,000, spread it on the plate with a spreader, and culture it overnight in a constant temperature incubator at 37°C. Pick a single colony and place it in an After 12 hours, transfer to 3ml of LB medium without anti-antibody. When the OD600 reaches 0.6, place it on ice for 15 minutes, centrifuge at 4000rpm / min at 4℃ for 3 minutes, and discard the supernatant Afterwards, the cells were reselected with 1ml of sterilized water, centrifuged at 4000rpm / min at 4°C for 3min, and repeated once, then resuspended with 100ul of 10% glycerol, quick-frozen in liquid nitrogen, and stored at -80°C.
[0053] 2. Preparation of Linearized Template
[0054] Use the genome of the BL21 Star (DE3) strain and the pTKS / CS plasmid as a template to carry out the PCR reaction according to...
Embodiment 2
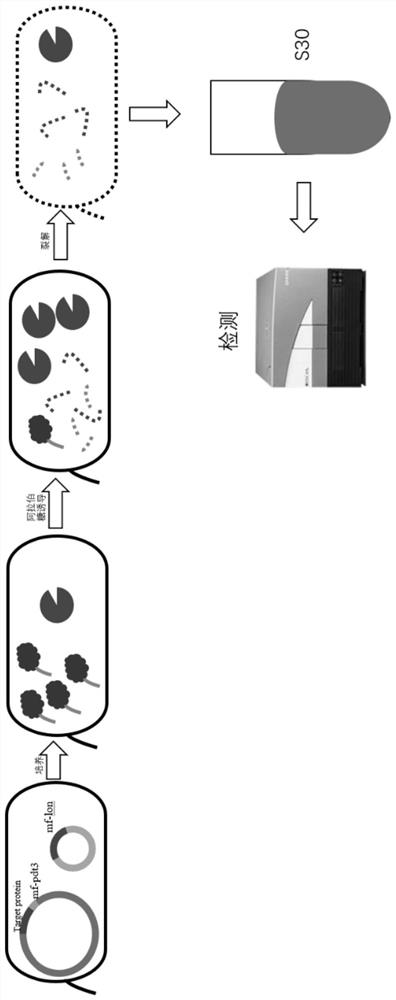
[0108] Example 2 Preparation of S30 Extraction and Inspection of RF1 and RF2 in Bacteria
[0109] 1. Preparation of electroporation competent and bacterial culture:
[0110] The correct sequenced BL21 Star(DE3)-SLA3B3 strain was prepared according to the above steps to prepare BL21 Star(DE3)-SLA3B3 electroporation competent, and then the pZA16mflon-pAzFRS plasmid was electrotransferred into BL21 Star(DE3)-SLA3B3, plated (Amp- LB plate), cultivate overnight at 37°C; pick a single colony from the plate, inoculate in 22mL of LB liquid medium, and culture overnight on a shaker at 37°C for 10 hours. A part of the bacteria needs to be frozen to preserve the species. Take 10 mL of the overnight bacterial solution, inoculate it in 1.0 L of S30 medium containing 1 mM arabinose and 1 mL of 100 mg / ml ampicillin sodium, and culture it on a shaker at 37°C for about 11 hours until OD 600 = About 1.7 to collect bacteria.
[0111] 2. Collect bacteria
[0112] ① Immediately after the culti...
Embodiment 3
[0132] The mensuration of embodiment 3 S30 extract liquid activity
[0133] 1. Preparation of GFP-WT, GFP-149TAG, GFP-149TGA and GFP-149TAA templates
[0134] Take the pET23a-GFP, pET23a-GFP149TAG, pET23a-GFP149TGA and pET23a-GFP149TAA strains preserved in the laboratory, streak on the plate, and then put them in the culture at 37°C for 14 hours. Pick a single colony and place it in 5ml of LB (100ug / ml Amp) at 220 rpm / min at 37°C for 14 hours, and then extract the plasmid according to the instructions of the kit. The plasmid was subjected to PCR reaction according to the following steps:
[0135] Table 14
[0136]
[0137] Reaction conditions:
[0138] Table 15
[0139]
[0140] After the PCR reaction, agarose gel electrophoresis was performed, and the gel was cut and recovered according to the steps described above. After measuring the concentration, store at -20°C.
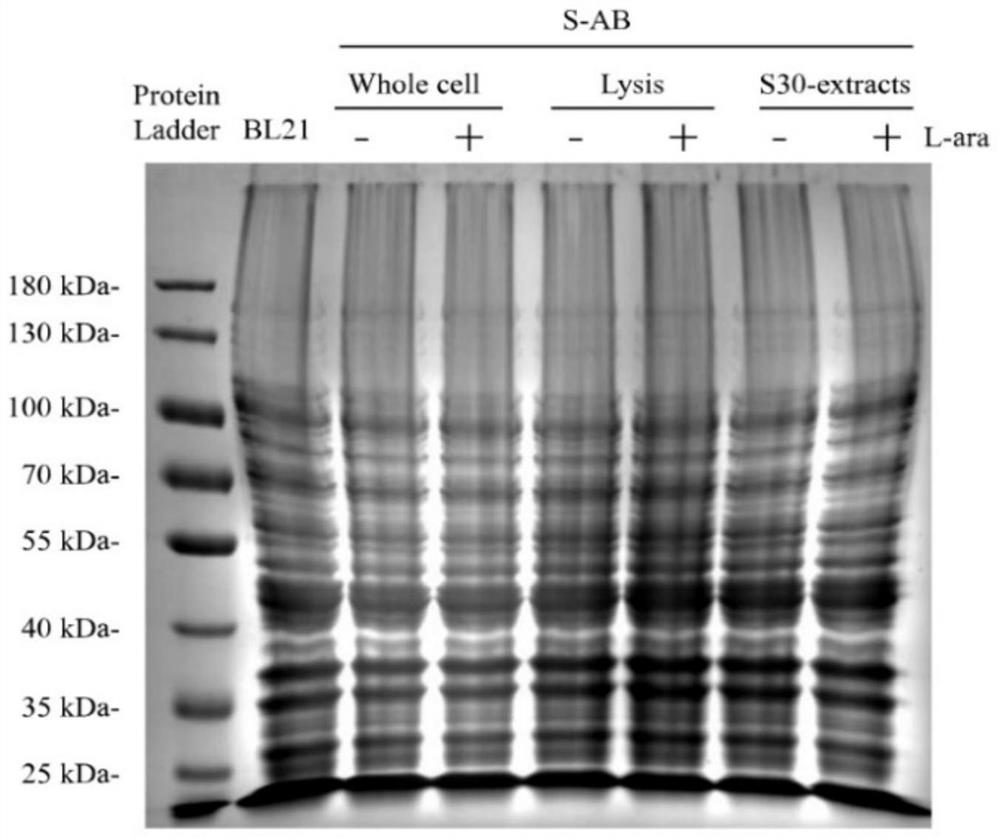
[0141] 2. Detection of activity of S30 extract
[0142] ①Preparation of 20×Amino Acid (20×AA): ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com