Phellinus mads-box transcription factor pbmads1 and its coding gene and application
A technology of transcription factors and coding genes, applied in the fields of application, genetic engineering, plant genetic improvement, etc., can solve the problems of long growth cycle and lack of research on growth and development regulation mechanism, so as to improve the growth rate and shorten the artificial cultivation period of Phellinus linteus. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] Embodiment 1, the cloning of Phellinus Phellinus PbMADS1 gene
[0021] Phellinus strain DL101 (the Phellinus DL101, Phellinus baumii DL101, is preserved in the China Type Culture Collection Center, address: China Type Culture Collection Center, Bayi Road, Hongshan District, Wuhan City, Hubei Province, China Type Culture Collection Center, postal code: 430072 , the preservation date is April 25, 2011, and the preservation number is: CCTCC No: M 2011137) was activated with PDA medium, cultured at 25°C for 8-10 days, collected Phellinus mycelia, and used the fast-type plant genomic DNA respectively The extraction kit and the RNAprep Pure plant total RNA extraction kit were used to extract the Phellinus mycelia DNA and RNA, and the RNA samples that passed the test were reversed according to the instructions of the Reverse Transcriptase M-MLV (RNase H) kit. Transcribed into cDNA. The Phellinus Phellinus DNA extracted and the cDNA obtained by reverse transcription were store...
Embodiment 2
[0026] Embodiment 2, the bioinformatics analysis of Phellinus Phellinus PbMADS1 gene
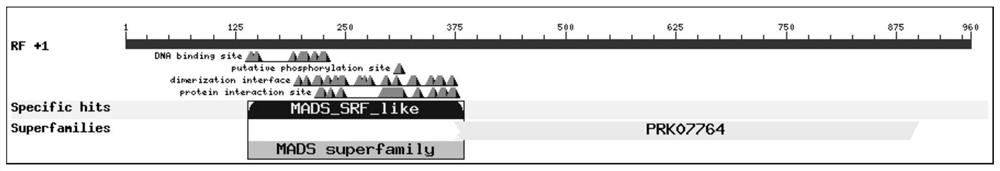
[0027] Using ProtParam tool software on the ExPASy server to analyze the molecular weight, isoelectric point and other physical and chemical properties of the amino acid sequence encoded by the PbMADS1 gene, the results show that its molecular weight is 32.71kDa, and the isoelectric point is 6.97; the prediction of the conserved domain of the protein shows that PbMADS1 protein belongs to MADS superfamily( figure 2 ); using the ProtScale software on the ExPASy server to analyze the affinity / hydrophobicity of amino acids, the number of hydrophobic amino acids in PbMADS1 protein is more than that of hydrophilic amino acids, which belongs to hydrophobic proteins; using online tools TMHMM Server v.2.0 and SignalP5.0server respectively The analysis of the transmembrane region and signal peptide of the protein shows that the PbMADS1 protein has no transmembrane structure and does not contain a sig...
Embodiment 3
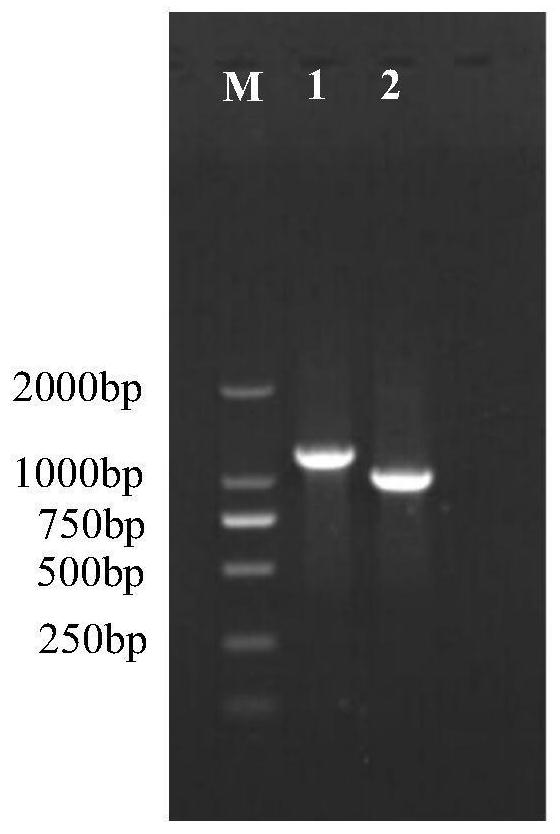
[0028] Example 3, Functional Identification of Phellinus Phellinus PbMADS1 Gene
[0029] The coding region of the PbMADS1 gene was cloned, using Phellinus cDNA as a template, primers were designed according to the coding region of PbMADS1, and BglⅡ restriction sites were introduced respectively,
[0030] FB: GGAagatctATGCAAGCAACTGACTCGCAAG
[0031] RB: GGAagatctTTACCCTCCTATACGTTGGCTGC,
[0032]The lowercase letters are the introduced BglⅡ restriction sites.
[0033] The pCAMBIA1301-gpd-gpd recombinant plasmid (the two CaMV35Spromoters on the pCAMBIA1301 vector had been replaced with the mushroom gpd promoter earlier) was digested with the restriction endonuclease BglⅡ and then gel-cut to recover, and the PbMADS1 The purified product of the gene coding region was ligated with T4 DNA ligase, transformed into Escherichia coli DH5α competent cells, positive clones were selected, and plasmids were extracted for sequencing.
[0034] The above-mentioned plasmids with correct seque...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com