Recombinant subunit vaccine of novel coronavirus South African mutant strain and application of recombinant subunit vaccine
A technology of coronavirus and mutant strains, applied in the direction of viruses, viral peptides, antiviral agents, etc., can solve the problems of poor protection of mutant strains in South Africa
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Embodiment 1, carrier construction
[0019] By chemical synthesis, Beijing Liuhe Huada Gene Technology Co., Ltd. synthesized the fusion protein-related gene sequence of the antigenic epitope of the new coronavirus South African mutant COVID-19 South African mutant vaccine. in,
[0020] The RBD nucleotide sequence of the novel coronavirus South African mutant strain COVID-19 South African mutant strain is as follows (SEQ ID NO: 1):
[0021]
[0022] The amino acid sequence of the encoded RBD is as follows (SEQ ID NO: 2):
[0023]
[0024] The encoded SD1 amino acid sequence is as follows (SEQ ID NO: 3):
[0025]
[0026] The nucleotide sequence of SD1 is as follows (SEQ ID NO: 4):
[0027]
[0028] The encoded hFc amino acid sequence is as follows (SEQ ID NO: 5):
[0029]
[0030] The nucleotide sequence of hFc is as follows (SEQ ID NO: 6):
[0031]
[0032] RBDSD1-hFc amino acid sequence (SEQ ID NO: 7)
[0033]
[0034] The nucleotide sequence...
Embodiment 2
[0038] Example two, cell transient
[0039] ①The day before transfection, according to 2.0×10 6 cells / mL density inoculation, the cell density can reach 4.0×10 on the second day of culture 6 cells / mL or so;
[0040] ② After counting the cells on the second day of culture, the cell viability is >95%, and the viable cell density is ≥4.0×
[0041] 10 6 cells / mL, can be used directly; if the cell density is lower than 4.0×10 6 cells / mL, the cells can be collected by centrifugation (800rpm, 5min), and the cells6 cells / mL density resuspended in Transpro CD01 medium;
[0042] ③According to the optimized transient process, prepare the DNA and PEI mixture;
[0043] ④ Add the mixed solution to the culture medium for cultivation;
[0044] ⑤After 18 hours of culture, it is recommended to add a final concentration of 2mM valproic acid (VPA) and an initial culture volume of 5%+0.5%DN feed 2+DN feed B2, which can further increase the density of viable cells and protein expression. If ...
Embodiment 3
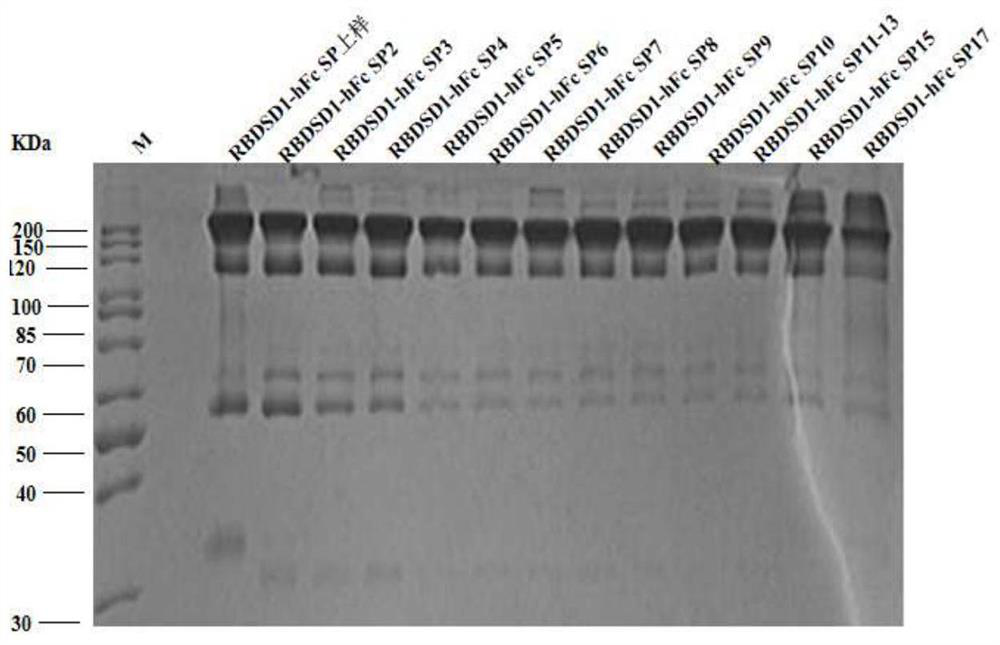
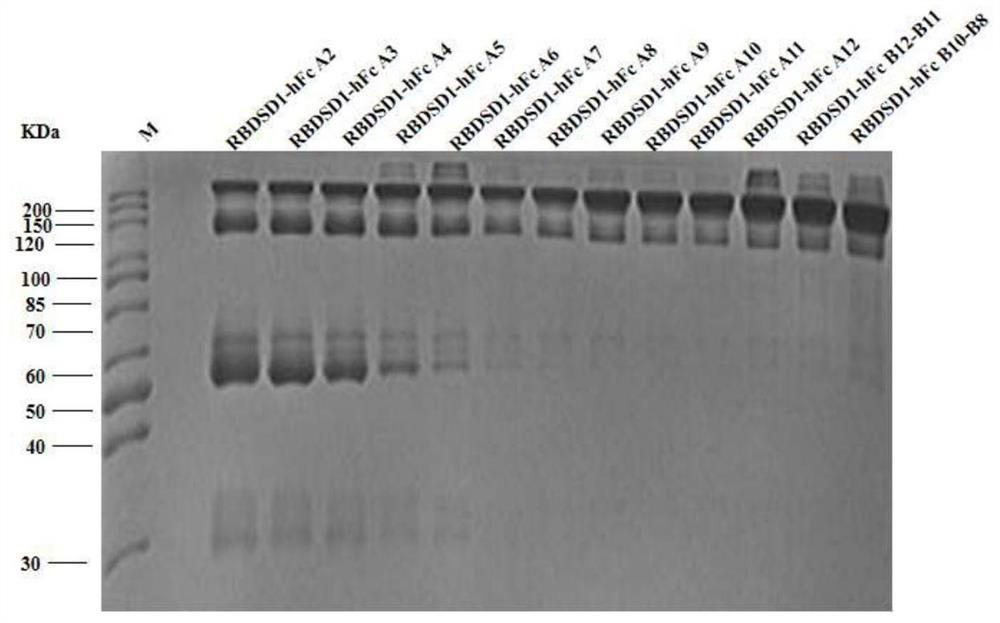
[0046] Embodiment three, protein purification
[0047] The protein extracted in Example 2 was purified by affinity chromatography and then ion exchange as follows.
[0048] 1. For the purification of RBDSD1-Fc, the following Protein A affinity chromatography method is adopted
[0049] One) the overall steps of the Protein A affinity chromatography method are as follows
[0050] Chromatography column: AT ProteinA Diamond affinity chromatography medium, column volume CV: 10ml, flow rate: 4-6ml / min, pressure limit: ≤0.3MPa.
[0051] Pretreatment: first rinse with 0.1M NaOH for at least 3 CVs, and then rinse with purified water for at least 10 CVs.
[0052] Equilibrium: Wash 2 to 3 CVs with washing buffer (0.1M HAc-NaAc, pH3.0), then fully equilibrate at least 10 CVs with binding buffer (0.02M PB, 0.5M NaCl, pH7.0) stand-by.
[0053] Sample loading: Take the filtered CHO suspension cell culture and use 5M NaCl to adjust the conductance to be consistent with the binding buffer,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com