Preparation method and application of nucleic acid library for low host background interference
A technology of background interference and nucleic acid library, applied in biochemical equipment and methods, chemical library, microbial measurement/inspection, etc., can solve problems such as deviation from target genes, lower detection accuracy, multi-species mixed sequencing pollution, etc., to reduce Nucleic acid interference, improving detection accuracy, improving the effect of nucleic acid targeted sequencing analysis technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
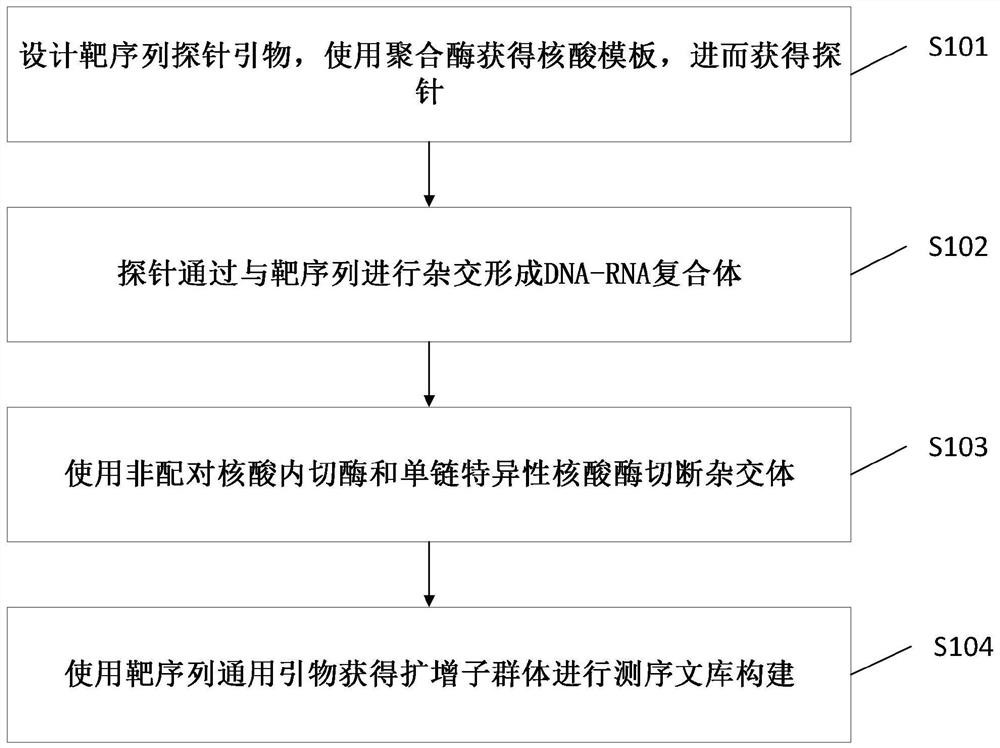
[0026] see figure 1 , figure 1 It is a schematic flowchart of a nucleic acid library preparation method for low host background interference provided by the embodiment of the present invention. Specifically, the nucleic acid library preparation method for low host background interference may include the following steps:
[0027] S101. Design target sequence probe primers, use polymerase to obtain nucleic acid templates, and then obtain probes;
[0028] In the embodiment of the present invention, amplification primers are designed for the target sequence to be sequenced. The primers can be used for the amplification of one or more amplicons. The primer: can be a single oligonucleotide, or a series of oligonucleotides, or a modified oligonucleotide. Non-targeted or targeted gene depletion using nucleic acids as probe source templates, which can be: linear DNA fragments, linear RNA fragments, nucleic acid fragments subcloned into circular vectors, or nucleic acid sequences fr...
Embodiment 1
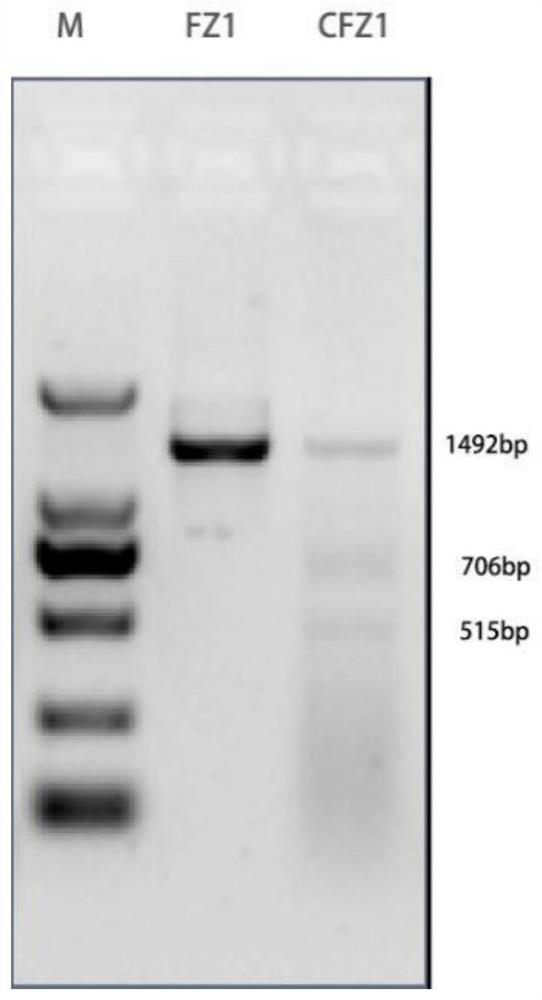
[0052] Example 1: Purification and Fragmentation of Sample Total DNA
[0053] The total DNA of the sample is purified by the CTAB method or the genomic DNA purification kit corresponding to the sample. The nucleic acid concentration of the purified total DNA was measured by a Qubit fluorometer or a Nano Drop spectrophotometer, and the integrity of the total DNA was detected by an Agilent2100 bioanalyzer.
Embodiment 2
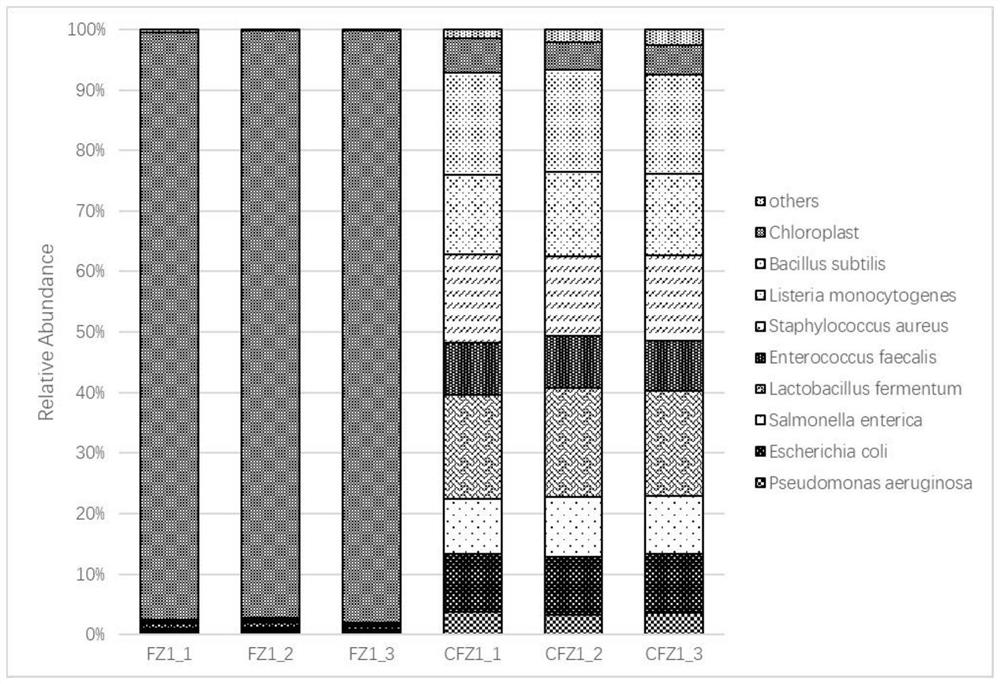
[0054] Example 2: Screening of target genes and preparation of probes
[0055] Taking the detection of endophytic prokaryotic microorganisms in Arabidopsis thaliana leaves as an example, the plant plastid and mitochondrial 16S gene (NC_000932.1) sequences were searched in the NCBI Nucleotide nucleic acid database, and the corresponding DNA was chemically synthesized according to the obtained Arabidopsis plastid and mitochondrial sequences. , and subcloned into pUC18 cloning vector, named pUC18-NC321. Use primers AT7_515F / U806R and U515F / AT7_806R to amplify respectively with pUC18-NC321 as template to obtain amplicons containing T7 promoter, then mix the two amplicons in equimolar amounts to obtain probe DNA template U4, the final concentration 50ng / μl.
[0056] Use U4 as a template and use T7 RNA polymerase to perform in vitro transcription to synthesize RNA probes. The reaction system is as follows:
[0057] components volume 10×Reaction Buffer 2μl A...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com