Ginkgo long-chain non-coding RNA Lnc2L and Lnc2S as well as vector and application thereof
A long-chain non-coding, Ginkgo biloba technology, applied in the field of plant genetic engineering, can solve problems such as few research reports and no relevant reports on lncRNAs.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Cloning of Lnc2L and Lnc2S
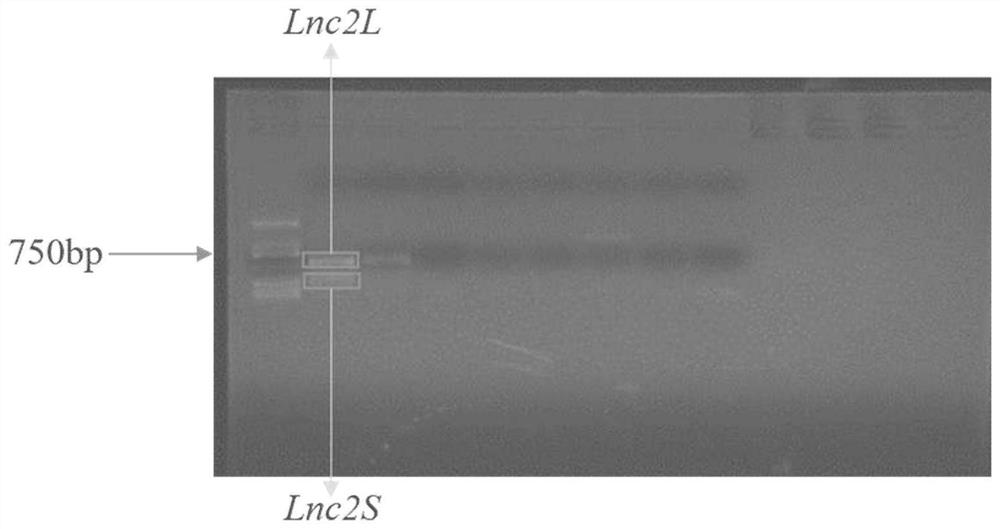
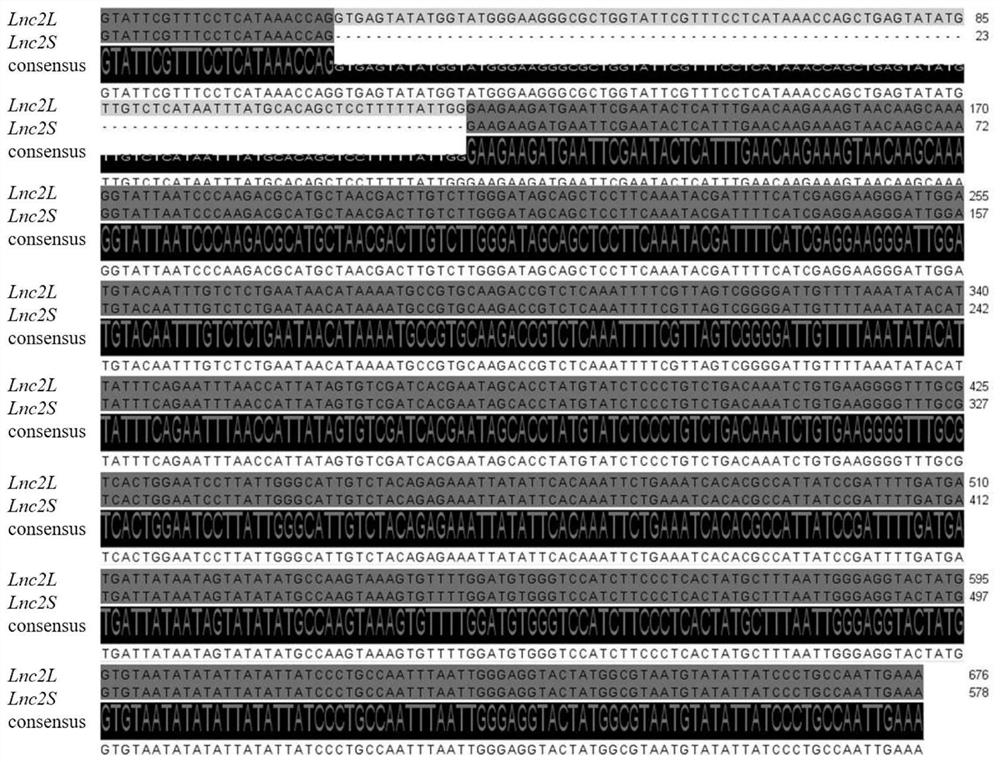
[0030] (1) Based on the Ginkgo lncRNA-seq data, an lncRNA was screened, and primers were artificially designed for this lncRNA using Primer Premier 5.0 software. Wherein the forward primer (F primer) is: 5'-GTATTCGTTTCCTCATAAACCAGG-3', and the reverse primer (R primer) is: 5'-TTTCAATTGGCAGGGATAATATA-3'. Two bands appeared during PCR amplification, which may be caused by variable splicing. The long sequence was named Lnc2L, and the short sequence was named Lnc2S( figure 1 ).
[0031] (2) Using the high-fidelity enzyme PrimeSTAR Max (Takara, Japan) for PCR amplification, the PCR system is as follows:
[0032]
[0033] Gently mix the above mixture, centrifuge briefly at low speed and place it in an ordinary PCR reaction instrument, set the following program:
[0034]
[0035] Gel running: Take out the gene amplification product in the PCR instrument, use the electrophoresis instrument to spot an appropriate amount of the product on a 1...
Embodiment 2
[0050] Construction of plant expression vectors for Lnc2L and Lnc2S
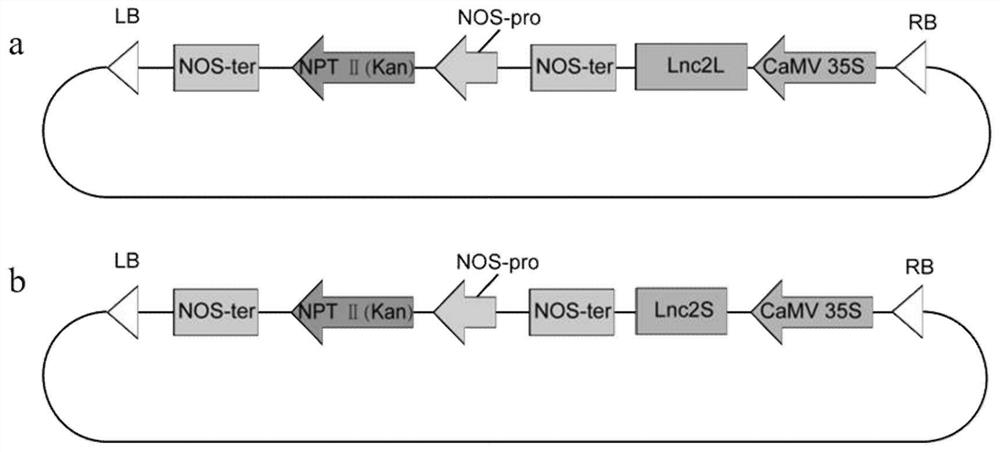
[0051] (1) This experiment uses TaKaRa QuickCut restriction enzyme (TaKaRa, Japan) to pRI 101-AN vector (TaKaRa, Japan, with promoter CAMV35S, strong terminator NOS-ter, NPTⅡ gene expression cassette, LB and RB Sequence) and the sequences of Lnc2L and Lnc2S were subjected to enzyme digestion reaction experiments, the specific reaction system is as follows:
[0052]
[0053] After the solutions in the system were mixed, they were centrifuged instantaneously, incubated in a 37°C water bath for 30 minutes, and then the enzyme digestion reaction was completed. The enzyme-cut bands were observed by agarose gel electrophoresis, and then the target gene and carrier fragments were cut and recovered for subsequent use. The carrier ligation reaction.
[0054] (2) Referring to the operation manual of TaKaRa T4 DNA Ligase (TaKaRa, Japan), connect the expression vector recovered after the double enzyme digestion reac...
Embodiment 3
[0061] Genetic transformation of Lnc2L and Lnc2S
[0062] 1. Ginkgo callus transformation
[0063] (1) Agrobacteria containing 35S::Lnc2L and Lnc2S vectors obtained in Example 2 were coated on LB plates respectively. After culturing, pick the single clone of Agrobacterium on the LB plate, inoculate it into LB liquid medium, and culture it at 28°C for 16h to OD 600 0.5-0.6;
[0064] (2) Put the bacterial solution into a centrifuge tube, centrifuge at 18°C, 3500rpm for 15min, and remove the supernatant;
[0065] (3) Add resuspension solution (100 mL MS liquid medium containing 100 μM acetosyringone) to the centrifuge tube to resuspend the bottom bacteria, and place it at room temperature for 2 h;
[0066] (4) Put the ginkgo callus fragments of the same size into the Agrobacterium resuspension, let it stand at room temperature and soak for 15min, then gently clip it out with tweezers, and suck off the resuspension liquid on the surface with sterile filter paper;
[0067] (5) ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com