Indel/SNP molecular marker related to flavor character of fresh edible soybeans and application of Indel/SNP molecular marker
A molecular marker and fragrance technology, applied in the field of agricultural biology, can solve the problems of high cost, volatile and low accuracy of content identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
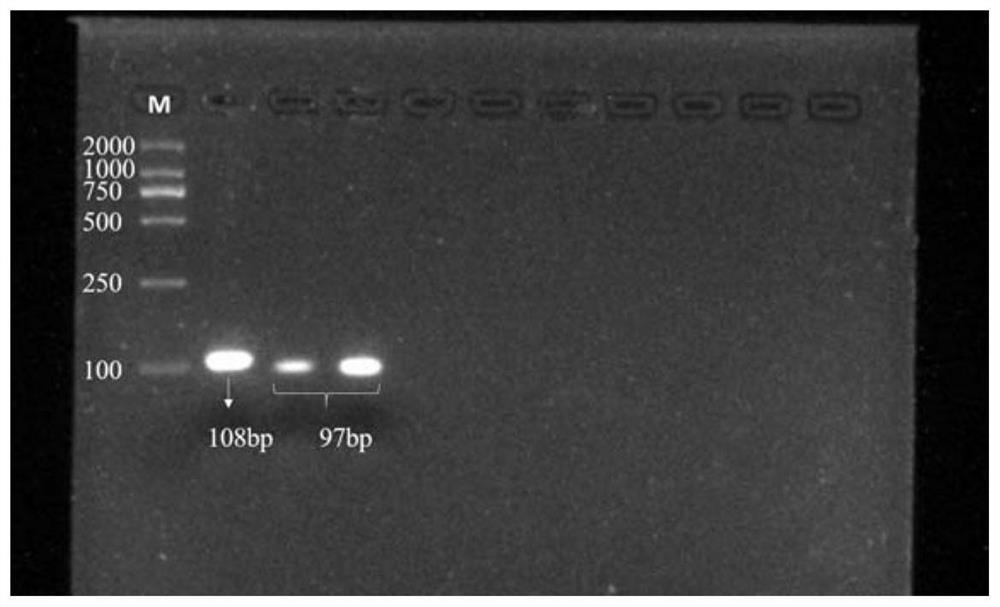
[0024] Example 1 HRM molecular marker detection method of fresh soybean aroma major gene GmBADH2
[0025] 1. Experimental equipment
[0026] Ultra-low temperature refrigerator (Thermo, the United States); high-speed refrigerated centrifuge (Eppendorf, Germany); gene mutation detector ( 480Ⅱ, Switzerland); adjustable micropipette (Eppendorf, Germany); chemiluminescent gel imaging system (G:BOX, China); electrophoresis instrument (BIO-RAD, USA).
[0027] 2. Soybean Genomic DNA Extraction
[0028] The leaves of soybean seedlings were taken, and the whole genome DNA of soybean was extracted with a plant genome DNA extraction kit (Solarbio, China). After extraction, 1% agarose gel electrophoresis is used to detect the quality of the DNA; a micro-nucleic acid concentration analyzer is used to measure the DNA concentration.
[0029] 3. Primer design
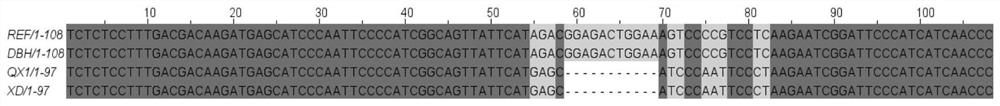
[0030] Specific primers were designed for the GmB ADH2 gene segment of chromosome 5 of the Glycine max Wm82.a2.v1 genome in the P...
Embodiment 2
[0039] Example 2 Verification test of the molecular marker in hybrid parents (Danbohei and Quxian No. 1)
[0040] 1. Test materials and methods
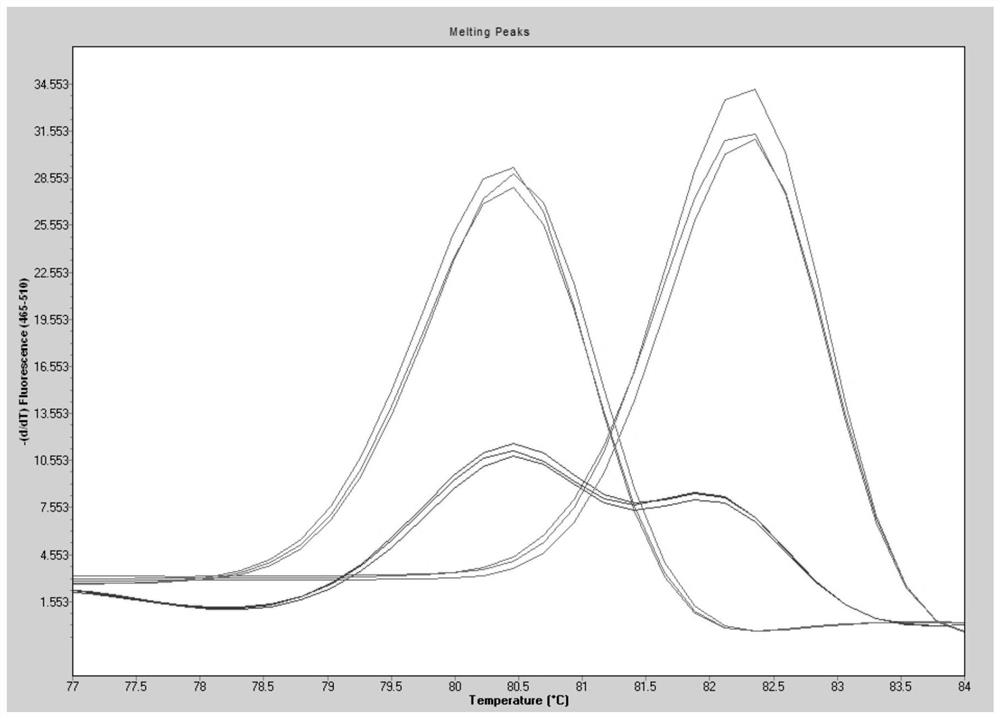
[0041]Danbahei comes from the Institute of Agricultural Sciences, Zhejiang Academy of Agricultural Sciences, and has no fragrance. Quxian No. 1 comes from Quzhou Academy of Agriculture and Forestry, and has fragrance. The genomic DNA of Danbahei and Quxian No. 1 was extracted, tested according to the steps in Example 1, and the genotype of the samples was judged according to the melting curve peak.
[0042] The method of GC-IMS was used to measure the 2AP content of Danbahei and Quxian No. 1, specifically: picking fresh pods, boiling the water and putting the soybeans in, taking them out 5-10 minutes after the water boiled again, removing the shells and putting them in the GC-IMS. IMS incubator. The detection conditions of the gas chromatography ion mobility spectrometer are: analysis time 20min, chromatographic column type FS-SE-5...
Embodiment 3
[0048] Example 3 Verification Test of Soybean Aroma-Related Indel / SNP Molecular Markers in Hybrid Populations
[0049] 1. Test materials and methods
[0050] A hybrid combination was prepared with Danbohei and Quxian 1 as parents to obtain hybrid seeds, the pseudo hybrids were eliminated in the F1 generation, F1 generation seeds were obtained, and F2 generation plants were obtained by field sowing; Genomic DNA was extracted from a single plant, and detected according to the steps in Example 1, and the genotype of the sample was judged according to the peak of the melting curve. The determination of 2AP content is the same as implementation case 2.
[0051] 2. Results and analysis
[0052] Of the 117 F2 individuals, 26 individuals had a melting temperature of 80°C and were homozygous recessive genotypes. The average content of 2 AP was 6.19 μg / g, which was similar to that of the parent Quxian No. 1; 32 individuals The melting temperature of the 59 individual plants was 80℃ a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com