Unlabeled proteomics detection method and device
A proteomics and detection method technology, applied in measurement devices, scientific instruments, biological testing, etc., can solve the problems of complex design, large sample size, and inability to continuously automatic sample injection, and achieve the effect of cost reduction and simple pretreatment.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
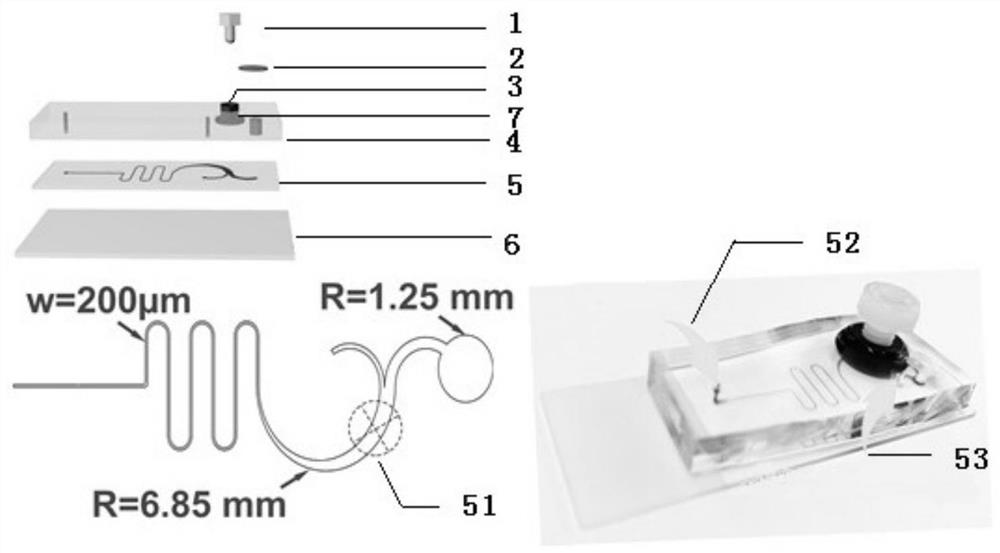
[0048] Example 1 Preparation of microfluidic system
[0049] (1) Pour PDMS and curing agent (Sylgard 184, Dow Corning) into a Petri dish on the surface of the silicon wafer at a ratio of 11:1, and bake at 85°C for 45 minutes.
[0050] (2) Use oxygen plasma to bond the nut to the PDMS solidified surface near the middle of the semicircular channel for 50 seconds.
[0051] (3) Pour PDMS and curing agent into a petri dish until the nuts are covered, and bake for 1 hour.
[0052] (4) Peel off the PDMS with nuts from the dish, and stick to the glass substrate after being treated with oxygen plasma for 50s.
[0053] (5) Punch holes at the inlet and outlet. The inlet and inner outlet adopt a punching machine with a diameter of 0.8mm, and the outer outlet adopts a punching machine with a diameter of 2.5mm.
[0054] (6) A filter membrane with a pore size of 20 μm is pasted on the top of the outlet.
[0055] (7) Treat the chip with 1% BSA for 1 h at room temperature to block the surf...
Embodiment 2293
[0056] Example 2 293T cell proteome sample preparation
[0057] (1) Prepare the target cells. After dilution, add the microfluidic system through the injection port and enter the sample pool.
[0058] (2) with 50mM NH 4 HCO 3 Chips were washed, replaced with PBS, and images of the cavity were taken through a Zeiss LSM 880, Germany microscope for accurate cell counts.
[0059] (3) The filter membrane is removed.
[0060] (4) The bolt is screwed into the nut to fix the cells in the exit mouth.
[0061] (5) Place the chip in an oven at 80°C for 30 minutes to denature cell proteins.
[0062] (6) Take 5 μL (0.1% DDM, 1mM TCEP, 2mM CAA dissolved in 50mM NH 4 HCO 3 ) to the outer export.
[0063] (7) The outer outlet was sealed with a sample bottle gasket and electrical tape, and incubated at 60° C. for 1 hour.
[0064] (8) Then add 5 μL of trypsin (total trypsin (w): protein (w) = 1:10) from the internal outlet, seal with the vial seal and electrical tape, and incubate overn...
Embodiment 3
[0067] Example 3 Separation, counting and proteome sample preparation of MCF7 cells
[0068] (1) The concentration of MCF7 cells was 5×10 4 cells / mL and 1×10 3 cells / mL and 5×10 respectively 4 The cells / mLWBCs are mixed, and 1mL / h of the microfluidic system is added through the injection port into the sample pool.
[0069] (2) Smaller cells and white blood cells flow out from the inner outlet, and MCF7 cells flow out from the outer outlet and pass through the filter membrane.
[0070] (3) with 50mM NH 4 HCO 3 Chips were washed, replaced with PBS, and images of the cavity were taken through a Zeiss LSM 880, Germany microscope for accurate cell counts.
[0071] (4) The filter membrane is removed.
[0072] (5) The bolts are screwed into the nuts to fix the cells in the exit mouth.
[0073] (6) Place the chip in an oven at 80°C for 30 minutes to denature cell proteins.
[0074] (7) Take 5 μL (0.1% DDM, 1 mM TCEP, 2 mM CAA dissolved in 50 mM NH4HCO3) and add it to the outer...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com