Neutralizing epitope peptide in RBD region of SARS-CoV-2 S protein and application of neutralizing epitope peptide
A sars-cov-2, sars-cov-2b technology, applied in the field of protein engineering, can solve the problems of inefficient expression of short peptides, lack of immunogenicity, neutralization of antigenic epitopes, and low immunogenicity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0136] Embodiment 1 recombinant antigen and antibody preparation
[0137] 1. Preparation of S1RBD
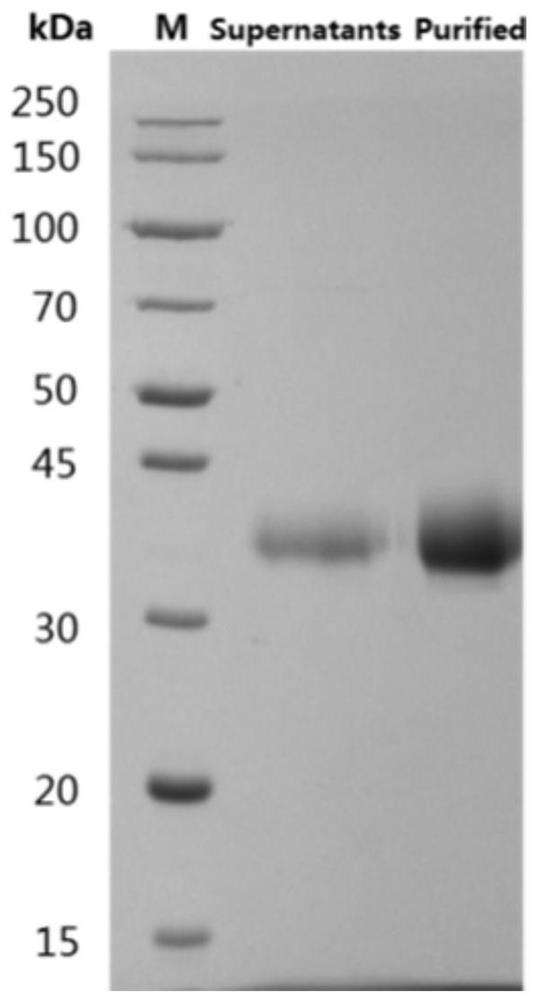
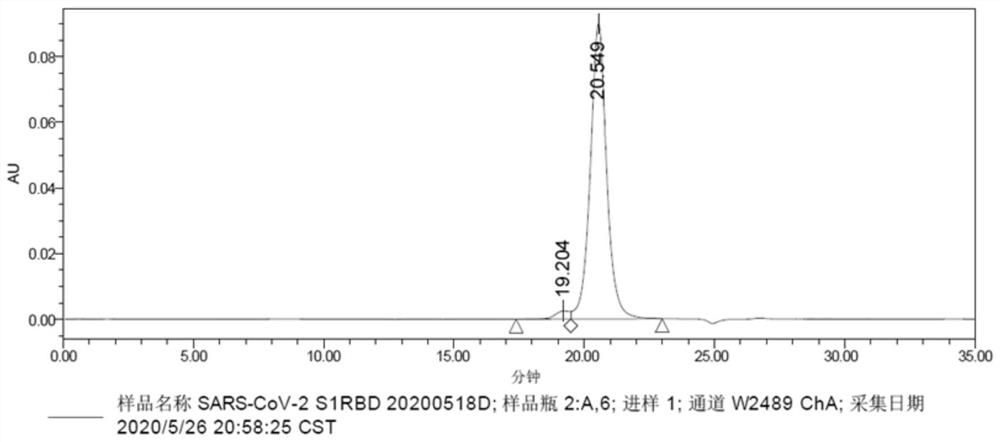
[0138] The SARS-CoV-2 S1RBD (QHD43416.1, 319-533aa) gene was cloned into a eukaryotic expression vector with a His tag at the N-terminal, and the S1RBD eukaryotic expression plasmid with a His tag at the N-terminal was constructed (recombinant RBD- The sequence of His is shown in SEQID NO: 23). The plasmid was transfected into HEK293 cells by transfection reagent 293fectin (Life Technologies, 12347-019, 1977248) for transient expression, and the supernatant was collected after 4 days, and HisTrap HP affinity chromatography column (GE, 17-5248-01 ) to purify the expression supernatant, then ultrafilter through an ultrafiltration concentration tube, replace the buffer with PBS, obtain the pure recombinant protein of SARS-CoV-2 S1RBD antigen, and analyze the purity by SDS-PAGE and HPLC, the results are as follows figure 1 shown.
[0139] 2. Preparation of MW01, MW06 and MW07-Fab...
Embodiment 2
[0143] Embodiment 2 Antigen-antibody complex preparation and analysis
[0144] 1. Preparation of MW01-Fab antigen-antibody complex
[0145] Mix N-terminal His tag SARS-CoV-2 S1RBD and MW01-Fab in 20mM Tris 150mM NaCl solution according to the ratio of SARS-CoV-2 S1RBD: MW01-Fab at 0.9:1, and place at room temperature for 2 hours to make MW01-Fab fully Combined, ultrafiltered through an ultrafiltration concentration tube (Cat: VS2002, Sartorius Stedim), concentrated to 20mg / ml, 0.3ml, and carried out SEC-HPLC and SDS-PAGE detection; according to the results, the antigen-antibody complex retention time Lower than SARS-CoV-2 S1RBD and MW01-Fab, the main peak is a single peak, suggesting that the complex sample meets the requirements of the crystallization experiment ( Figure 6 ).
[0146] 2. Preparation of MW05-Fab antigen-antibody complex
[0147] Mix SARS-CoV-2 S1RBD and MW05-Fab in 20mM Tris 150mM NaCl solution according to the molar ratio SARS-CoV-2 S1RBD: MW05-Fab = 1:2....
Embodiment 3
[0152]Example 3 MW05 Antigen Antibody Crystal Preparation and Diffraction Data Collection and Structure Analysis
[0153] 1. Crystal Index and Diffraction
[0154] Crystals of MW05 antigen-antibody complexes were grown using Crystal Screen1, CrystalScreen2, Index, PEGRx1, PEGRx2 from HAMPTON, and WIZARD I / II kits from Emerald Biosystems. Using the sitting drop method, under the conditions of 0.2M sodium citrate, 21% PEG3350, and 0.02M urea, crystallized for 3 days, and finally obtained crystals with better diffraction, such as Figure 10 shown. The crystal was collected at the Shanghai Synchrotron Radiation Facility Diffraction data of MW317 FAB and covid-19Spike in MW317-PD1 complex structure (PDB ID: 6JJP) using PHASER software RBD -The spike protein RBD in the hACE2 complex structure (PDB ID: 6LZG) is a molecular replacement model, and the complex structure of MW05 is analyzed by the molecular replacement method.
[0155] 2. Analysis of key amino acids involved in S1RB...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com