Broad-spectrum neutralizing antibody for resisting novel coronavirus and application of broad-spectrum neutralizing antibody
A coronavirus and antibody technology, applied in the field of immunity, can solve problems such as drug resistance and reduced neutralization activity, and achieve the effect of avoiding biosafety risks
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
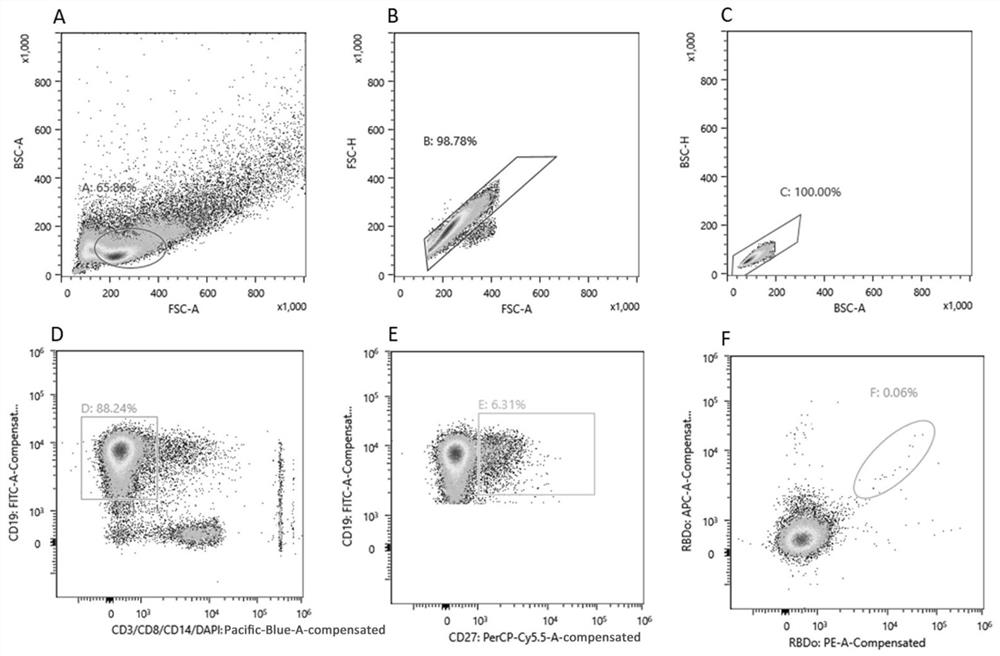
[0078] Example 1. Preparation of anti-novel coronavirus monoclonal antibodies
[0079] 1. Detection of plasma binding activity
[0080] 1. Sample: plasma of volunteers immunized twice with novel coronavirus inactivated vaccine (BBIBP-CorV).
[0081] 2. Detection of binding activity: The binding titers of 28 samples to the RBD protein of the novel coronavirus Omicron mutant strain (B.1.1.529) were detected. Methods as below:
[0082] (1) Coating: Dilute the above RBD protein with PBS to 0.5 µg / ml, add 100 µl / well to a 96-well ELISA plate, and coat overnight at 4°C.
[0083] (2) Blocking: discard the coating solution, add 200 µl blocking solution (PBST+3%BSA) to each well, and block at 37°C for 1 h.
[0084] (3) Washing: The blocking solution was discarded and washed three times with PBST.
[0085] (4) Sample: each plasma sample was diluted 100 times, serially diluted 3 times, and a negative control was added, and incubated at 37°C for 2 h.
[0086] (5) Washing: The sample ...
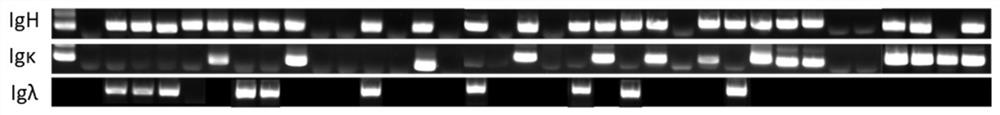
Embodiment 2
[0173] Example 2. Preparation of monoclonal antibodies
[0174] 1. Antibody expression in different cell lines
[0175] 1. Antibody expression in 293T cell system
[0176] Transient transfection of successfully paired antibody heavy and light chain gene expression vectors into 293T cells:
[0177] (1) 24 hours before transfection in a 12-well plate, each well was plated with 5 × 10 5 293T cells.
[0178] (2) Observe the degree of confluence of cells on the day of transfection, preferably 70% to 80%.
[0179] (3) Take 1 μg of each plasmid DNA extracted by the plasmid mini-extraction kit (Tiangen, DP103) and dilute it with 60 μl of Opti-MEM medium.
[0180] (4) Dilute 4 μl Lipofectamine® 2000 Reagent with 60 μl Opti-MEM medium.
[0181] (5) Mix the above-diluted DNA and Lipofectamine® 2000 at a volume of 1:1 to obtain a mixed solution, and incubate at room temperature for 5 min.
[0182] (6) Gently add 120 μl of the mixture to 293T cells, 5% CO 2 Incubate at 37°C.
[01...
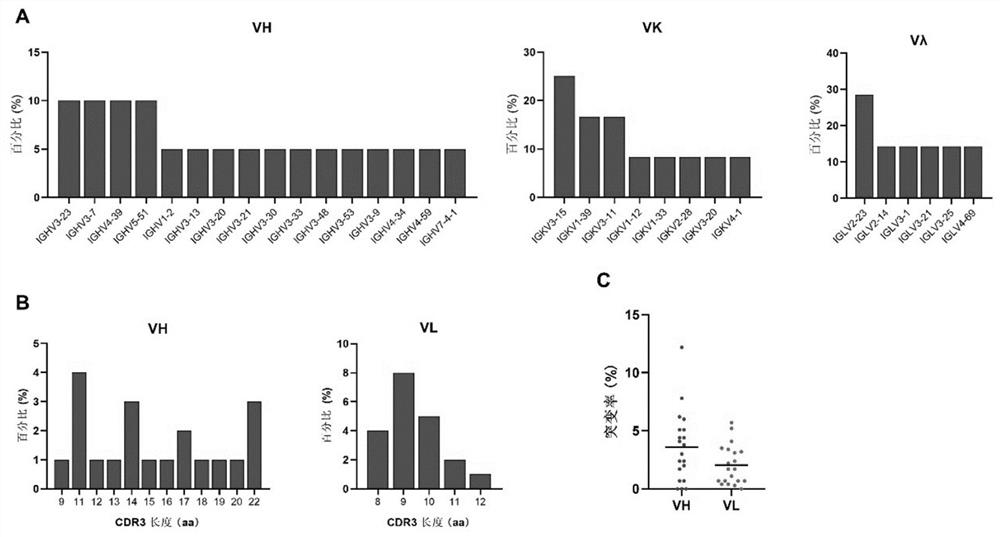
Embodiment 3
[0242] Example 3. Antibody 10-5B Affinity and Neutralizing Activity Analysis
[0243] 1. Affinity analysis of monoclonal antibodies
[0244] 1. BLI detects the binding ability of monoclonal antibody 10-5B to 2019-nCoV RBD
[0245] (1) Ligand coupling: Biotin-labeled 10 µg / ml wild-type RBD protein (acrobiosystems, SPD-C82E9) and Omicron (Omicron) were mutated on Octet RED 384, a high-throughput intermolecular interaction instrument. The RBD protein of the strain (Sino Biological, 40592-V49H7-B) was coupled to the SA sensor (SARTORIUS, 18-0009).
[0246] (2) Antibody 10-5B was serially diluted 2-fold in PBS from 160 nM in 6 gradients.
[0247] (3) Set the injection analysis from low concentration to high concentration according to Table 5 below.
[0248] Table 5. Low to High Injection Settings
[0249]
[0250] (4) Data analysis: The following Table 6 settings are performed in the analysis software, and curve fitting is performed.
[0251] Table 6 Analysis software setti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com