Nucleic acid aptamer for specifically recognizing soybean antigen beta-conglycinin
A technology of conglycinin and nucleic acid aptamer, which can be used in DNA/RNA fragments, material testing products, biological testing, etc., and can solve the problems of long time consumption, unsuitable for application, and unsuitable for large-scale testing.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0080] Example 1: Artificial synthesis to obtain specific nucleic acid aptamer Apt1
[0081] Entrusted Shanghai Sangon Bioengineering Co., Ltd. to synthesize the specific nucleic acid aptamer Apt1 of β-conglycinin and its complementary sequence, wherein the sequence of Apt1 is as follows:
[0082] AACACGACGGCCCTGTGTCCATCACAGTGTTGTGCATGTGGGC (SEQ ID NO: 1).
Embodiment 2
[0083] Example 2: Establishment of a fluorescent sensor method for the detection of β-conglycinin by Apt1 aptamer
[0084] 1) Construction of aptamer fluorescence system
[0085] First buffered with Tris HCl (50 mM Tris-HCl, 100 mM NaCl, 10 mM MgCl 2 , pH7.5) dilute Apt 1 aptamer and corresponding complementary sequence to a concentration of 10 μM, then add 50 μL of 10 μM Apt 1 aptamer and 50 μL of 10 μM complementary sequence cDNAs to a 1.5 mL centrifuge tube, vortex After rotating and oscillating evenly, put it in a water bath, and heat it at 95 °C for 10 min to release its own spiral. Finally, the nucleic acid mixture to be heated was cooled to room temperature for renaturation, and diluted 100 times with double distilled water to obtain 50 nM double-stranded DNA (dsDNA) synthesized by Apt1 aptamer and its complementary sequence, which was stored in a refrigerator at 4°C overnight for ready for use.
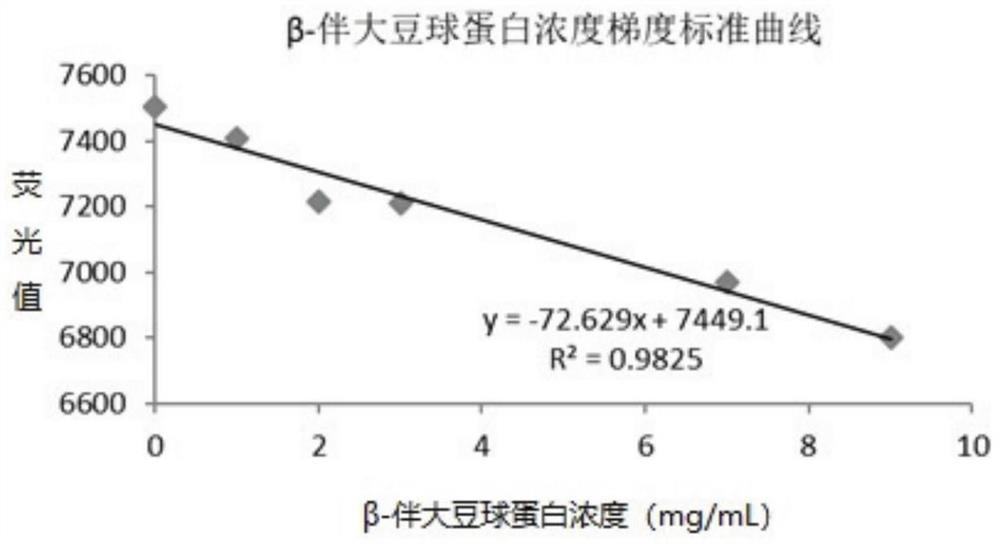
[0086] 2) Establishment of fluorescence detection method
[0087] 15 ...
Embodiment 3
[0096] Example 3: Detection of β-conglycinin in fermented soybean meal products with higher drying temperature
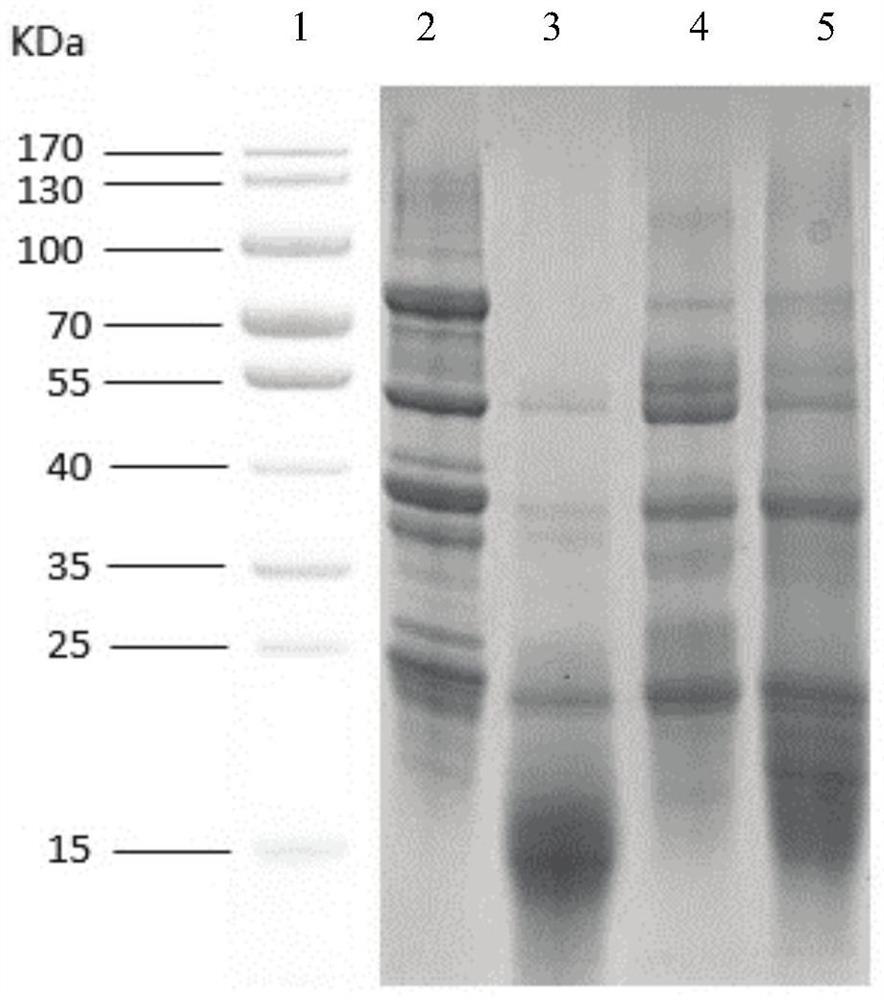
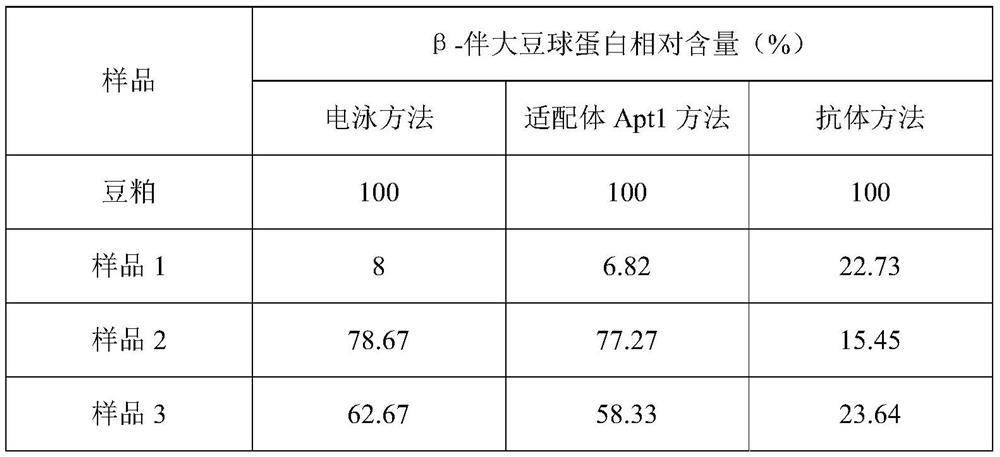
[0097] Sample 1, sample 2 and sample 3 are all fermented soybean meal products in the domestic market, and unfermented soybean meal is used as a control. The β-conglycinin content of the samples was detected by electrophoresis method, aptamer Apt1 method and antibody method (commercially available Elisa kit) respectively, and the test results of soybean meal were used as the standard 100%, and other test results were divided by the results of soybean meal. Degradation ratio value of β-conglycinin. in,
[0098] The electrophoresis method process is as follows: take 3 grams of the crushed sample with a size of 60 mesh in a triangular flask, add an appropriate amount of sample extract, mix well, then shake at 37°C for 2 hours, centrifuge at 10,000 rpm for 10 minutes, and take the supernatant. Add the loading buffer, take a boiling water bath for 5 minutes, and perfor...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com