DNA walker for SARS-CoV-2 detection and preparation method thereof
A sars-cov-2 and walker technology, applied in the field of biological detection, can solve problems such as low efficiency, achieve reliable methods, reduce the need for complex equipment, and achieve high sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
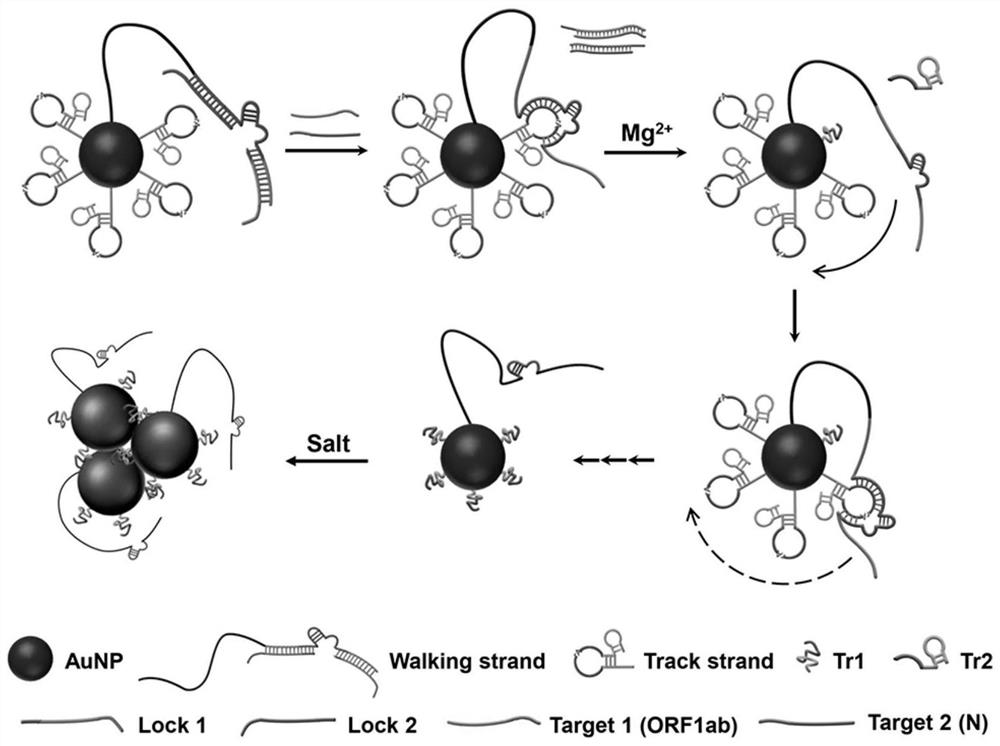
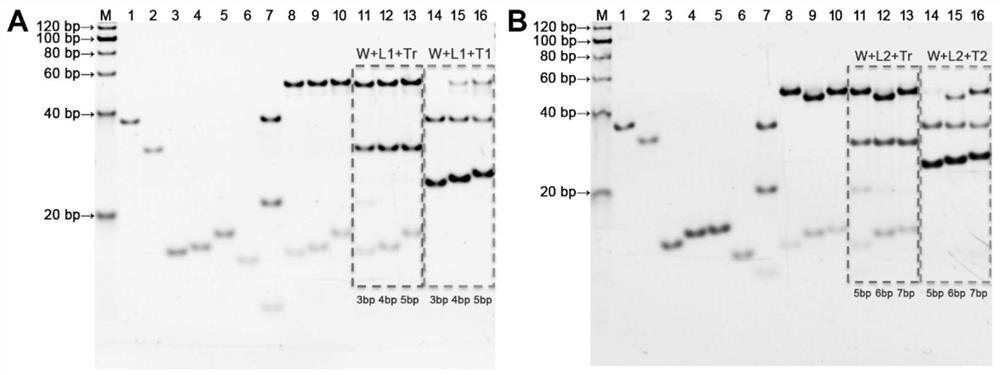
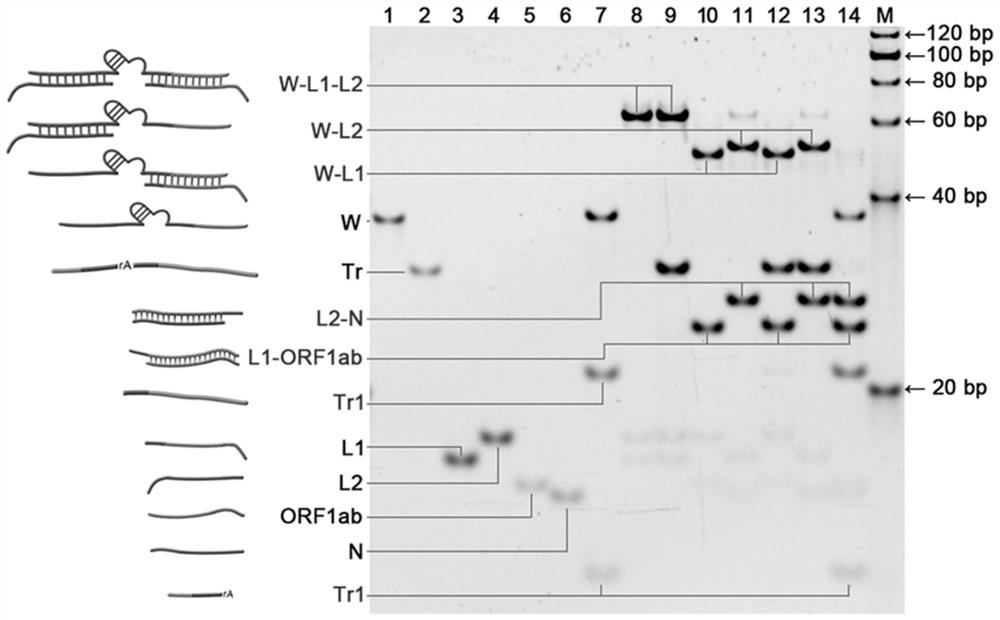
[0064] The present invention selects two RNA gene fragments ORF1ab and N from the open reading frame and nucleocapsid as the target chain T1 and the target chain according to the SARS-CoV-2 virus genome information (NCBI reference sequence: NC_045512.2) that has been disclosed in the NCBI database. T2, then according to the target strands T1, T2, the walking chain W, 3bp-locked strand L1, 4bp-locked strand L1, 5bp-locked strand L1, 5bp-locked strand L2, 6bp-locked strand L2, 7bp-locked strand L2 were designed , orbital chain Tr, the specific sequence is shown in Table 1. It was then synthesized and purified by Sangon Bioengineering Co., Ltd. (Shanghai, China).
[0065] The locked chain consists of a target recognition domain and a closed domain with different lengths.
Embodiment 2
[0067] 1. Preparation and storage of DNA and RNA solutions
[0068]Concentrated DNA stocks of the sequences described in Table 1 were prepared in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and diluted to working concentration with HEPES buffer (10 mM HEPES, 300 mM NaCl, pH 7.0).
[0069] Concentration method: Dissolve the nucleic acid sequence in microliters of TE buffer solution marked on the tube, and measure the concentration of the concentrated solution by ultraviolet absorption spectroscopy.
[0070] 2. Selection of locking chain L1 and locking chain L2
[0071] Mix 1.44 μL of 10 mM walking chain W and 1.50 μL of 10 mM locked chain (nbp-L1, nbp-L2) in annealing buffer (25 mM Tris-acetate, 200 mM NaCl, pH 8.0), respectively. The volume was 26.12 μL, the mixture was heated to 95°C for 2 min, and then cooled to 25°C to obtain an annealing mixture.
[0072] Combine 2.88 μL of 10 mM orbital chain Tr and 1 μL of 600 mM MgCl 2 Add to the annealing mixture, incubate at 25°C...
Embodiment 3
[0086] 1. The preparation method of gold nanoparticles is as follows:
[0087] Prepare 10 mL of sodium citrate aqueous solution with a concentration of 38.8 mM, and prepare 100 mL of HAuCl with a concentration of 1 mM 4 Aqueous solution; HAuCl 4 The aqueous solution was heated to boiling, then the aqueous sodium citrate solution was rapidly added to the HAuCl with vigorous stirring 4 The aqueous solution was heated and boiled for 10 min, stirred for 15 min after the reaction was completed, cooled to 25°C, and filtered through a 0.22 μm filter to obtain gold nanoparticles (AuNPs).
[0088] A drop of gold nanoparticle solution was added to the carbon-coated copper grid and dried at room temperature to prepare a sample for TEM characterization. Characterization was performed on a JSM-6700F transmission electron microscope with an accelerating voltage of 200 kV.
[0089] The UV-Vis absorption spectrum of AuNPs in this example is as follows: Figure 4 A, transmission electron m...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com