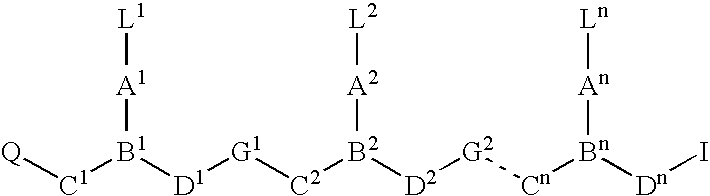
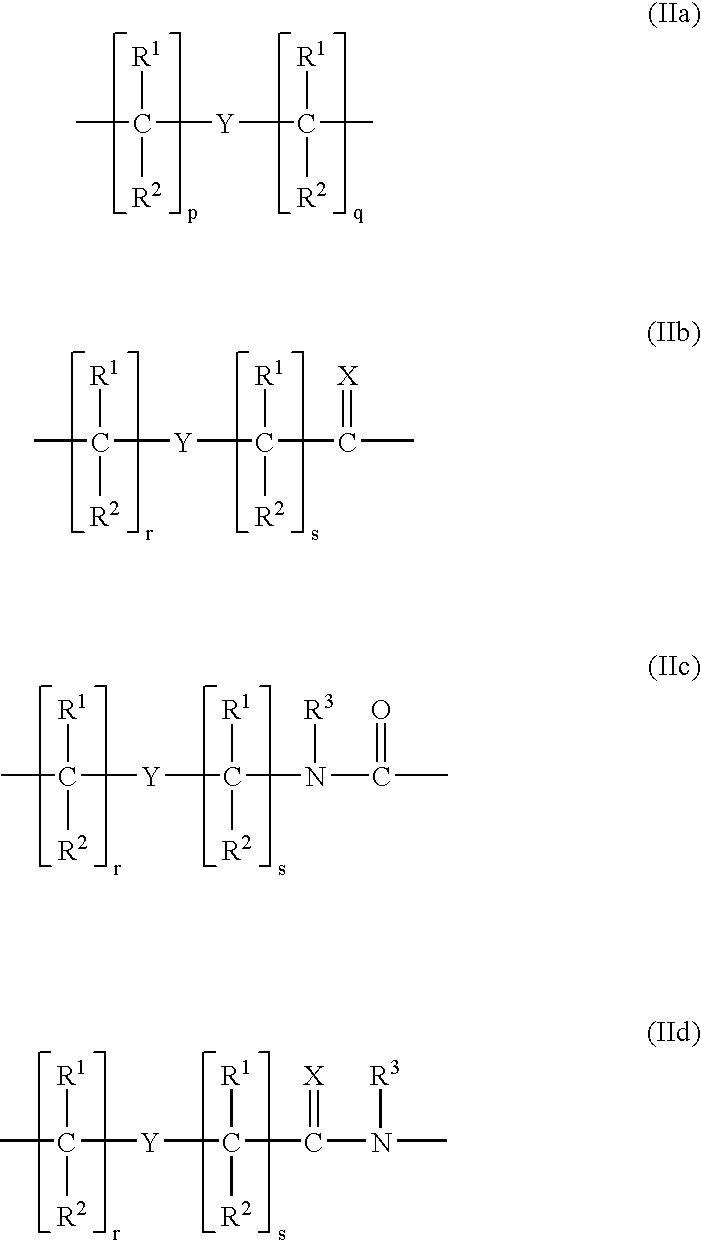
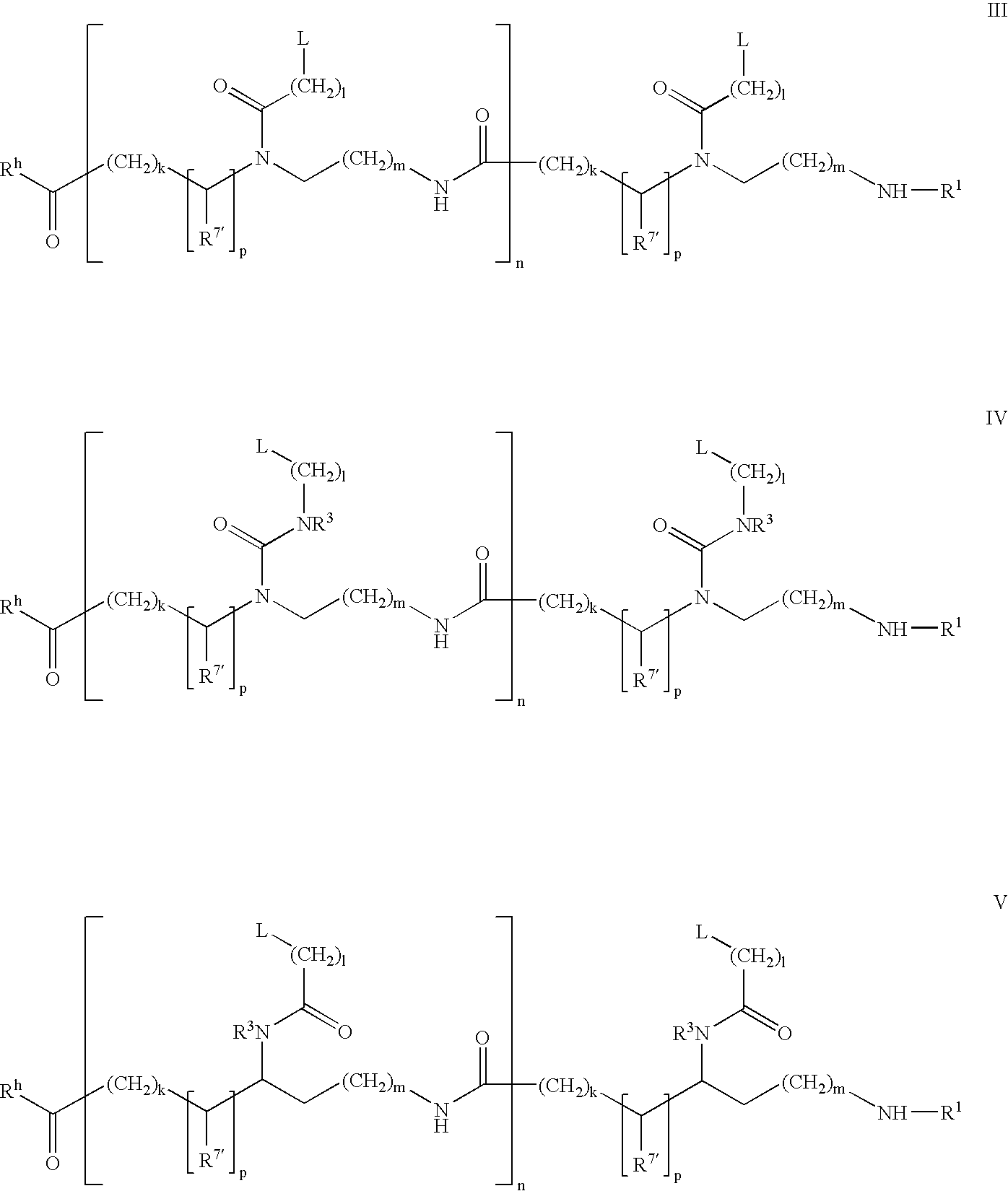
Higher order structure and binding of peptide nucleic acids
a peptide nucleic acid, higher order technology, applied in the direction of peptides, peptide/protein ingredients, organic chemistry, etc., can solve the problems of unpractical unmodified oligonucleotides, unphysiologically high ionic strength and low ph, and generally the formation of triple helixes, so as to facilitate the identification of suitable pna targets, stable strand displacement complexes, and certain effects on transcription
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 2
[0158] Site-Specific S.sub.1 Nuclease Digestion of the pT10 Plasmid Linearized with Cfr10I Restriction Enzyme and Complexed with PNA (a Double-Stranded Target)
[0159] PNA H-T.sub.10-LysNH.sub.2 (SEQ ID NO:2) was synthesized as described in Example 1. A pT10 plasmid was prepared from the pUC19 plasmid by inserting the dA.sub.10 / dT10 (SEQ ID NO:3) sequence into the BamH1 site of the polylinker as per the method of Egholm, M et al., J.A.C.S. 1992 114:1895-1897. The pT10 plasmid was linearized with Cfr10I restriction enzyme in the unique site. To form complex with the PNA, about 0.1 .mu.g of the linearized plasmid was incubated with 2 o.u. / ml of PNA in 3 .mu.l of the TE buffer (10 mM Tris-HCl; 1 mM EDTA, pH 7.4) at 37.degree. C. To perform the S.sub.1 nuclease reaction, 10 .mu.l of Na-Acetate buffer (33 mM NaAc; 50 mM NaCl; 10 mM ZnSO.sub.4; 0.5% of glycerol, pH 4.6) and 145 units of S.sub.1 nuclease (Sigma) were added and incubated for various time periods at room temperature. The react...
example 3
[0165] Site-Specific S.sub.1 Nuclease Digestion of the pT9C Plasmid Linearized by CfrlOl Restriction Enzyme and Complexed with PNA H-T.sub.5CT.sub.4-LysNH.sub.2
[0166] This example illustrates the use of one molecule of PNA hybridized to a target DNA to define a restriction site.
[0167] PNA 1, 2 and 3 were synthesized as Example 1 and the results are shown in FIG. 2. A pT9C plasmid carried the insert:
3 5'-A.sub.5GA.sub.4GTCGACA.sub.5GA.sub.4-3' (SEQ ID NO: 30) 3'-T.sub.5CT.sub.4CAGCTGT.sub.5CT.sub.4-5',
[0168] cloned in the Sal1 site of the pUC19 polylinker. The pT9C-5 plasmid carried the single insert A.sub.5GA.sub.4 / CT.sub.5 cloned in the same site. The PNA-DNA complexes were prepared as described in Example 2 with the only difference being the duration of the incubation was 2 hours. Digestion by 30 units of the S.sub.1 nuclease was performed in the same buffer as described in Example 2 with two exceptions, lanes 2 and 4. In lane 2, 15 units of the enzyme were used, whereas in lane 4...
example 4
[0170] Site Specific S.sub.1 Nuclease Cleavage of the Plasmids pT9C-5, pT9CT9C and pT9CA9GKS (Linearized with Sca1) Targeted by PNA T.sub.4CT.sub.5-LysNH.sub.2
[0171] This example illustrates the use of two molecules of PNA bound to opposite strands of a DNA target. A further plasmid pT9CA9GKS was produced by cloning the insert GTCGACA.sub.5GA.sub.4GTCGACT.sub.4CT.sub.5-GTCGAC (SEQ ID NO:32) into pUC19 at the Sal I site and linearizing with Sca I restriction enzyme. The linearized plasmid has two hybridization sites for PNA H-T.sub.4CT.sub.5-Lys NH.sub.2 (SEQ ID NO:8) on opposite strands spaced by six base pairs. The protocol of Example 3 was use except the samples were treated using 1 U / .mu.l of S.sub.1 and 15 min incubation at 37.degree. C. The results are shown in FIG. 3 Lanes 1-4: pT9C-5; lanes 5-8: pT9CT9C; lanes 9-12: pT9CA9GKS. Lanes 1, 5 & 9: no PNA; lanes 2, 6 & 10: 50 .mu.M; lanes 3, 7, 11: 500 uM; lanes 4, 8, 12: 5 mM.
EXAMPLE 5
[0172] Use of PNA / Nuclease to Cut Selectively ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| body weight | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com