Recombinant DNA constructs and methods for controlling gene expression
a technology of dna and constructs, applied in the field of molecular constructs and methods for controlling gene expression, can solve the problem of low efficiency of anti-sense gene suppression, and achieve the effects of suppressing the expression of a target rna, reducing damage to a plant, and reducing the accumulation of mature mirna
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
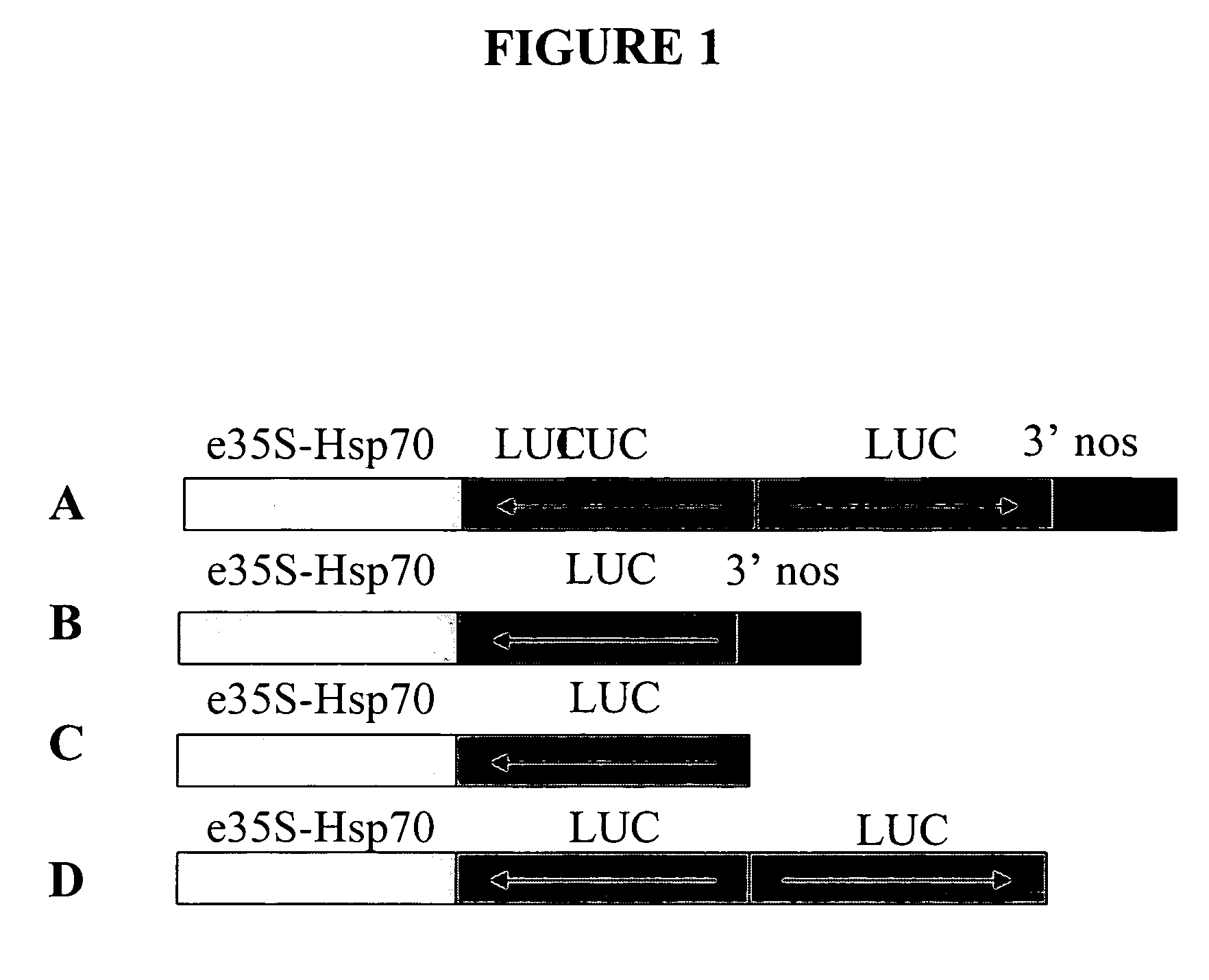
[0266] This example illustrates the construction and use of vectors designed for double-stranded RNAi suppression or for anti-sense suppression of a luciferase gene. The gene suppression experiments used were similar to a dual luciferase assay described by Horstmann et al. (2004) BMC Biotechnol., 4:13, which is incorporated by reference herein. A prior art vector, “vector 1A”, designed for double-stranded RNAi suppression of a luciferase gene was constructed as depicted in FIG. 1A with an RNAi transcription unit with a polyadenylation site including (a) a chimeric promoter including an enhanced CaMV35S promoter linked to an enhancer element (an intron from heat shock protein 70 of Zea mays, Pe35S-Hsp intron), (b) an inverted repeat of DNA coding for firefly luciferase (LUC) with anti-sense oriented DNA followed by a sense oriented DNA, and (c) a 3′UTR DNA from Agrobacterium tumefaciens nopaline synthase gene (3′NOS) which provides a polyadenylation (polyA) site. Elements of the plas...
example 2
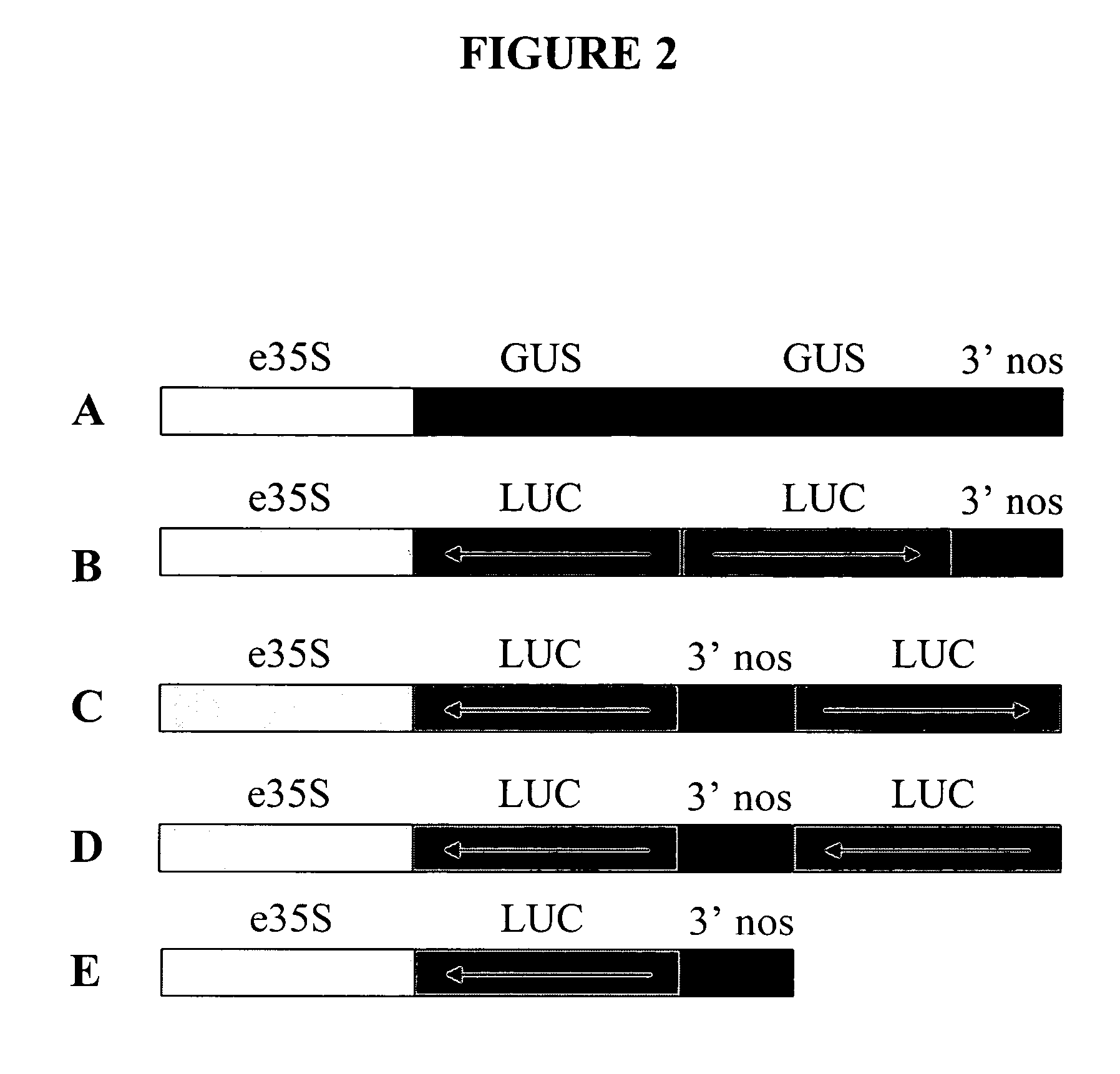
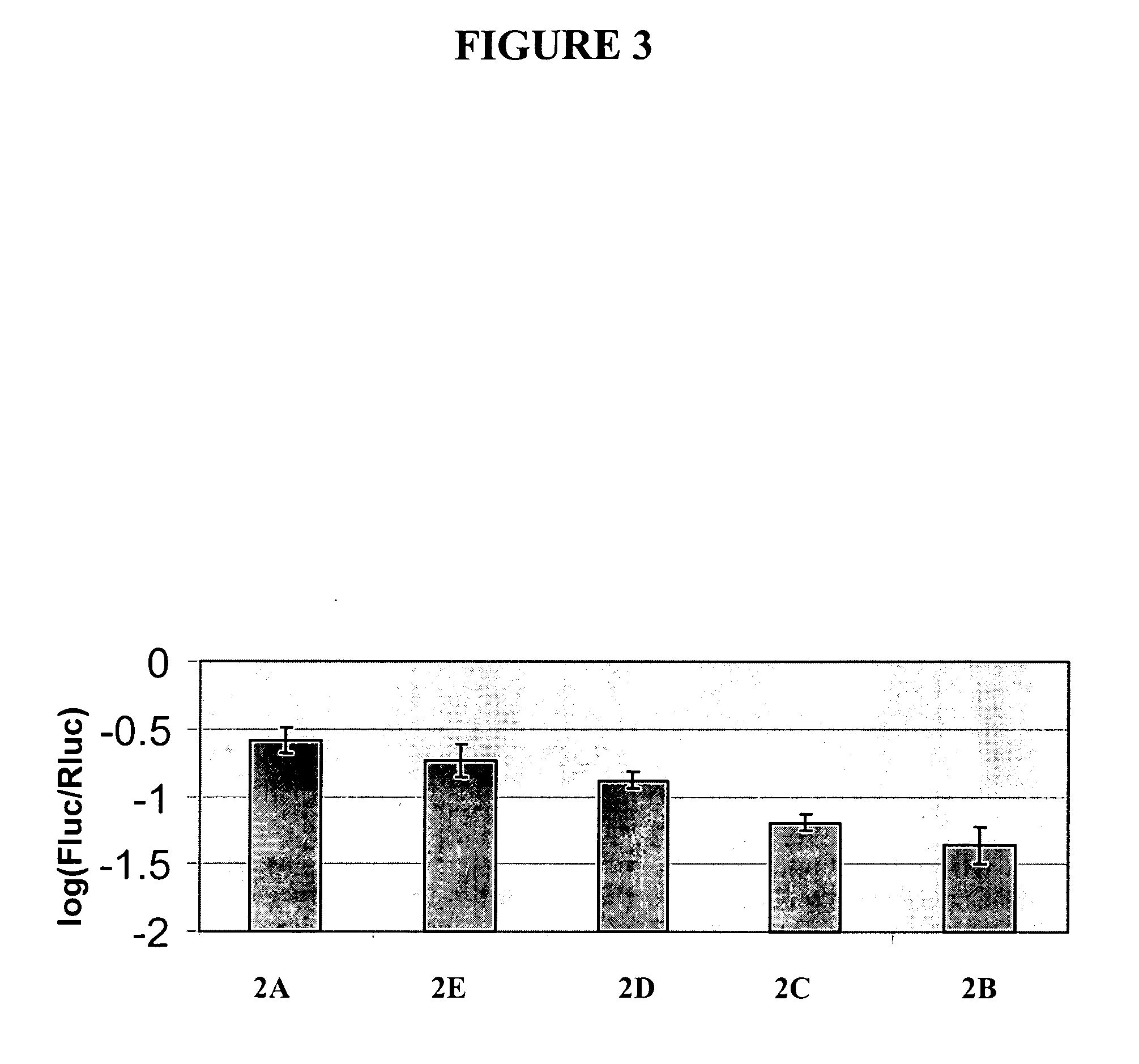
[0271] This example further illustrates the construction and use of vectors designed for double-stranded RNAi suppression or for anti-sense suppression of a luciferase gene. The gene suppression experiments used were similar to a dual luciferase assay described by Horstmann et al. (2004) BMC Biotechnol., 4:13. The vectors illustrated in FIG. 2 were constructed. Vector 2A (FIG. 2A), a control vector not encoding anti-sense or double-stranded RNA for the target gene (firefly luciferase), consisted of (a) the CaMV e35S-Hsp 70 intron chimeric promoter as described in Example 1 and Table 1, (b) an inverted repeat of DNA coding for beta-glucuronidase (GUS) (uidA) with anti-sense oriented DNA followed by a sense oriented DNA, and (c) a 3′UTR DNA from Agrobacterium tumefaciens nopaline synthase gene (3′NOS) as described in Example 1 and Table 1, which provides a polyadenylation (polyA) site. Vector 2B (FIG. 2B), a prior art vector designed for double-stranded RNAi suppression of a luciferas...
example 3
[0273] This example describes transformation of a crop plant (maize) with an enhanced anti-sense construct. A plasmid for binary vector Agrobacterium-mediated transformation of maize is constructed including the elements shown in FIG. 4. Specifically, the plasmid includes an nptlI gene as an antibiotic selectable marker and a recombinant DNA construct for enhanced anti-sense gene suppression, consisting of a CaMV35S promoter operably linked to transcribable DNA consisting of about 300 base pairs of a green fluorescent protein (gfp) gene in an anti-sense orientation, wherein a functional polyadenylation site is absent in this transcribable DNA. The plasmid also includes left T-DNA border (LB) and right T-DNA border (RB) elements. A control plasmid for RNAi suppression of green fluorescent protein (GFP) is constructed by adding to the enhanced anti-sense construct shown in FIG. 4 a repeat of the gfp DNA in the sense orientation followed by a 3′ NOS element including a functional polya...
PUM
| Property | Measurement | Unit |
|---|---|---|
| optical density | aaaaa | aaaaa |
| biotic stress tolerance | aaaaa | aaaaa |
| resistance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com