Molecular antigen array
a technology of molecular antigen and array, which is applied in the field of molecular biology, virology, immunology and medicine, can solve the problems of not being able the risk of serious complications of a reversion to virulence and infection by the ‘vaccine’ organism, and the inability to carry out the antigen array, etc., and achieves high efficiency.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Insertion of the JUN Amphiphatic Helix Domain Within E2
[0266] In the vector pTE5′2J (Hahn et al., Proc. Natl. Acad. Sci. USA 89: 2679-2683, (1992)), MluI and a BstEII restriction enzyme sites were introduced between codons 71 (Gln) and 74 (Thr) of the structural protein E2 coding sequence, resulting in vector pTE5′2JBM. Introduction of these restriction enzymes sites was done by PCR using the following oligonucleotides:
Oligo 1:E2insBstEII / BssHII:(SEQ. ID NO: 1)5′-ggggACGCGTGCAGCAggtaaccaccgTTAAAGAAGGCACC-3′Oligo 2:E2insMluIStuI:(SEQ ID NO: 2)5′-cggtggttaccTGCTGCACGCGTTGCTTAAGCGACATGTAGCGG-3′Oligo 3:E2insStuI:(SEQ ID NO: 3)5′-CCATGAGGCCTACGATACCC-3′Oligo4:E2insBssHII:(SEQ ID NO: 4)5′-GGCACTCACGGCGCGCTTTACAGGC-3′
[0267] For the PCR reaction, 100 pmol of each oligo was used with 5 ng of the template DNA in a 100 μl reaction mixture containing 4 units of Taq or Pwo polymerase, 0.1 mM dNTPs and 1.5 mM MgSO4. All DNA concentrations were determined photometrically using the GeneQuant app...
example 2
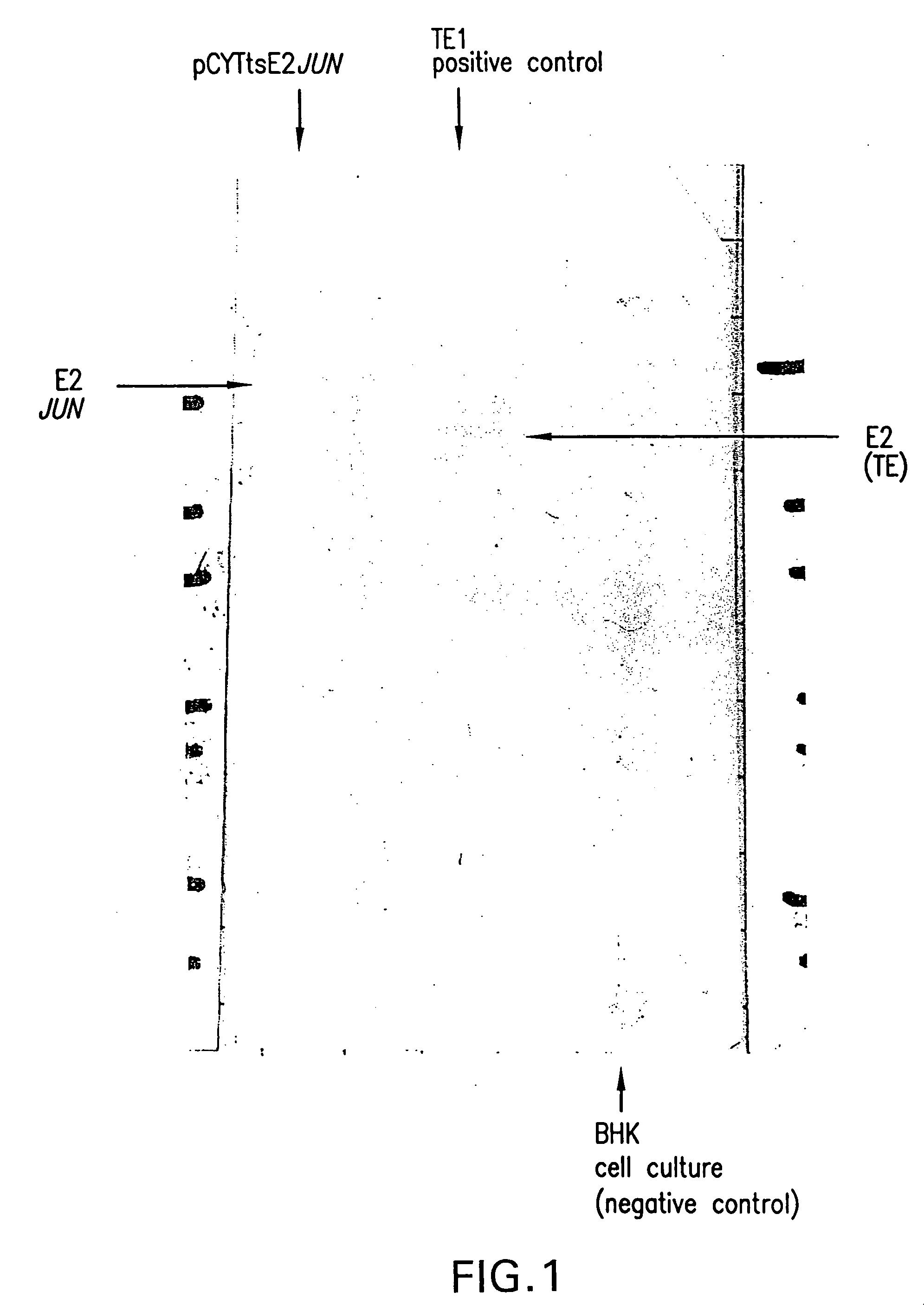
Production of Viral Particles Containing E2-JUN Using the pCYTts System
[0274] The structural proteins were PCR amplified using pTE5′2J: E2JUN as template and the oligonucleotides XbalStruct (ctatcaTCTAGAATGAATAGAGGATTCTTTAAC (SEQ ID NO:12)) and StructBsp1201 (tcgaatGGGCCCTCATCTTCGTGTGCTAGTCAG (SEQ ID NO:87)). For the PCR 100 pmol of each loligo was used and 5 ng of the template DNA was used in the 100 μl reaction mixture, containing 4 units of Tac or Pwo polymerase, 0.1 mM dNTPs and 1.5 mM MgSO4. All DNA concentrations were determined photometrically using the GeneQuant apparatus (Pharmacia). The polymerase was added directly before starting the PCR reaction (starting point was 95° C.). The temperature cycles were as follows: 95° C. for 3 minutes, followed by 5 cycles of 92° C. (30 seconds), 54° C. (35 seconds), 72° C. (270 seconds) and followed by 25 cycles of 92° C. (30 seconds), 63° C. (35 seconds), 72° C. (270 seconds. The PCR product was gel purified and digested with the rest...
example 3
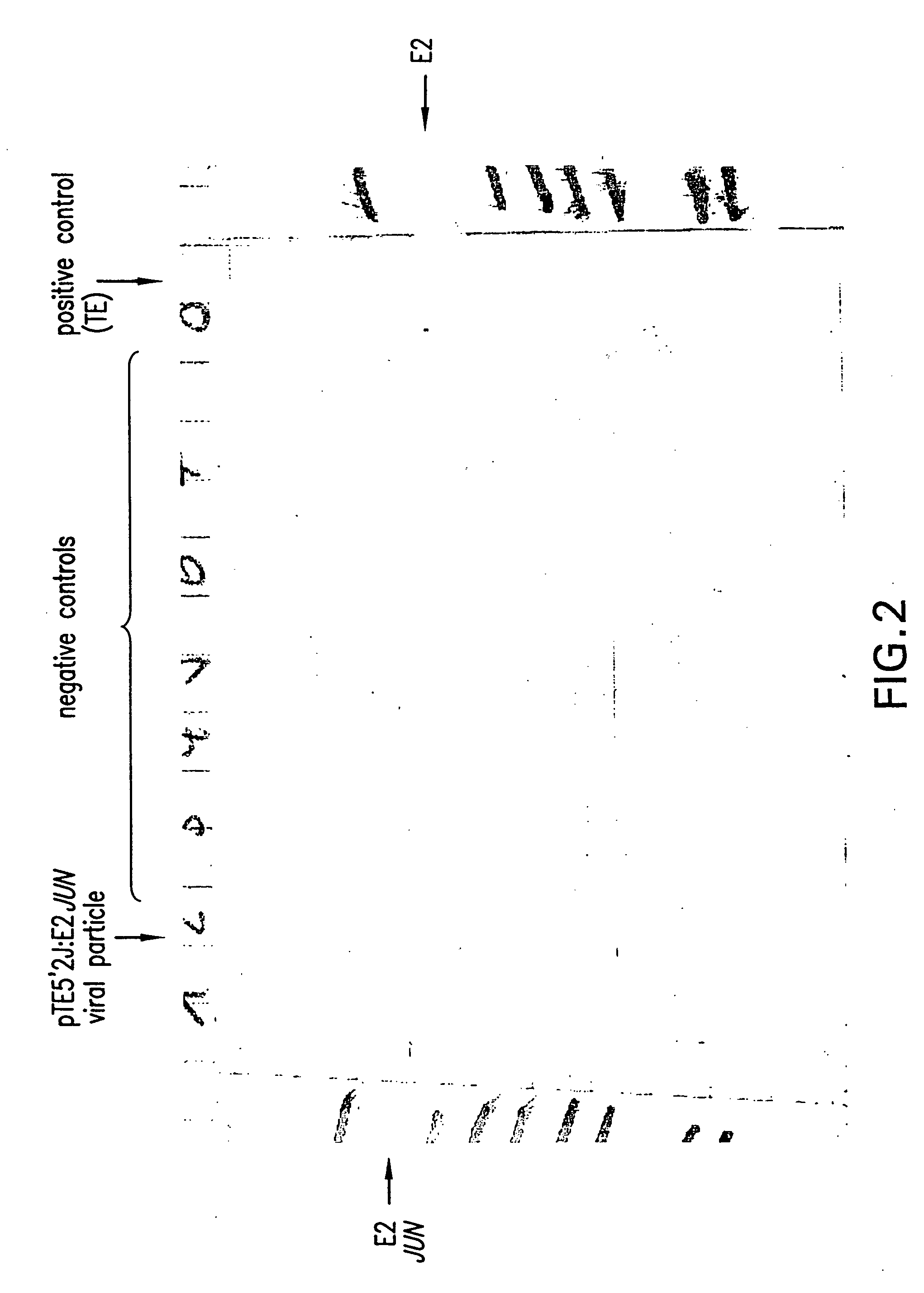
Production of Viral Particles Containing E2-JUN Using the pTE5′2JE2: JUN Vector
[0279] RNase-free vector (1.0 μg) was linerarized by PvuI digestion. Subsequently, in vitro transcription was carried out using an SP6 in vitro transcription kit (InvitroscripCAP by InvitroGen, Invitrogen BV, NV Leek, Netherlands). The resulting 5′-capped mRNA was analyzed on a reducing agarose-gel.
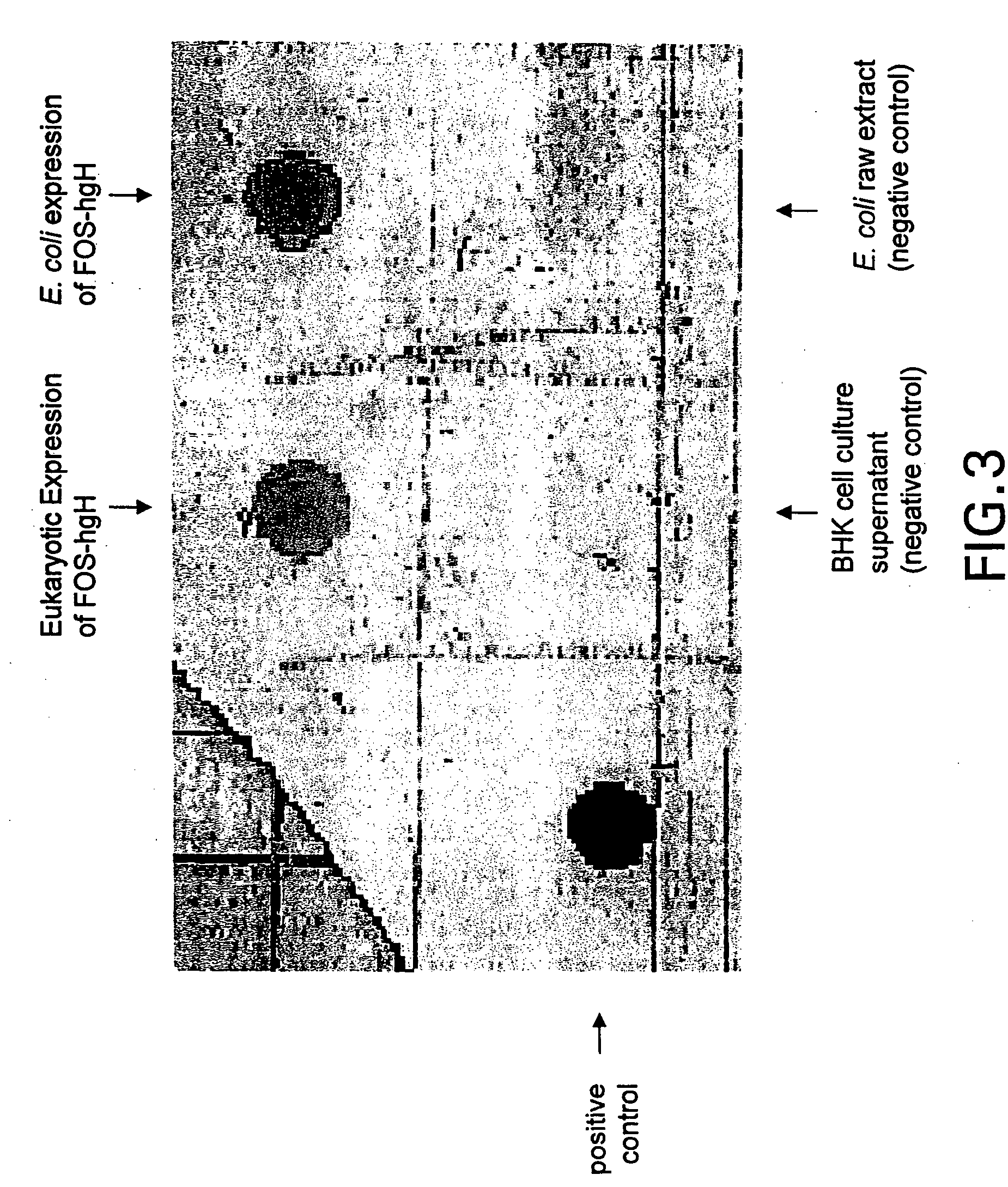
[0280] In vitro transcribed mRNA (5 μg) was electroporated into BHK 21 cells (ATCC: CCL10) according to Invitrogen's manual (Sindbis Expression system, Invitrogen BV, Netherlands). After 10 hours incubation at 37° C., the FCS containing medium was exchanged by HP-1 medium without FCS, followed by an additional incubation at 37° C. for 10 hours. The supernatant was harvested and analyzed by Western blot analysis for production of viral particles exactly as described in Example 2.
[0281] The obtained result was identical to the one obtained with pCYTtsE2: JUN as shown in FIG. 2.
PUM
| Property | Measurement | Unit |
|---|---|---|
| optical density | aaaaa | aaaaa |
| temperatures | aaaaa | aaaaa |
| temperatures | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com