Methods and systems for direct sequencing of single DNA molecules
a single dna and dna technology, applied in the field of methods and systems for direct sequencing of single dna molecules, can solve the problem of elusive $1000 genome technology, and achieve the effect of rapid dna sequencing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Design of Labeled Phi-29 DNA Polymerase
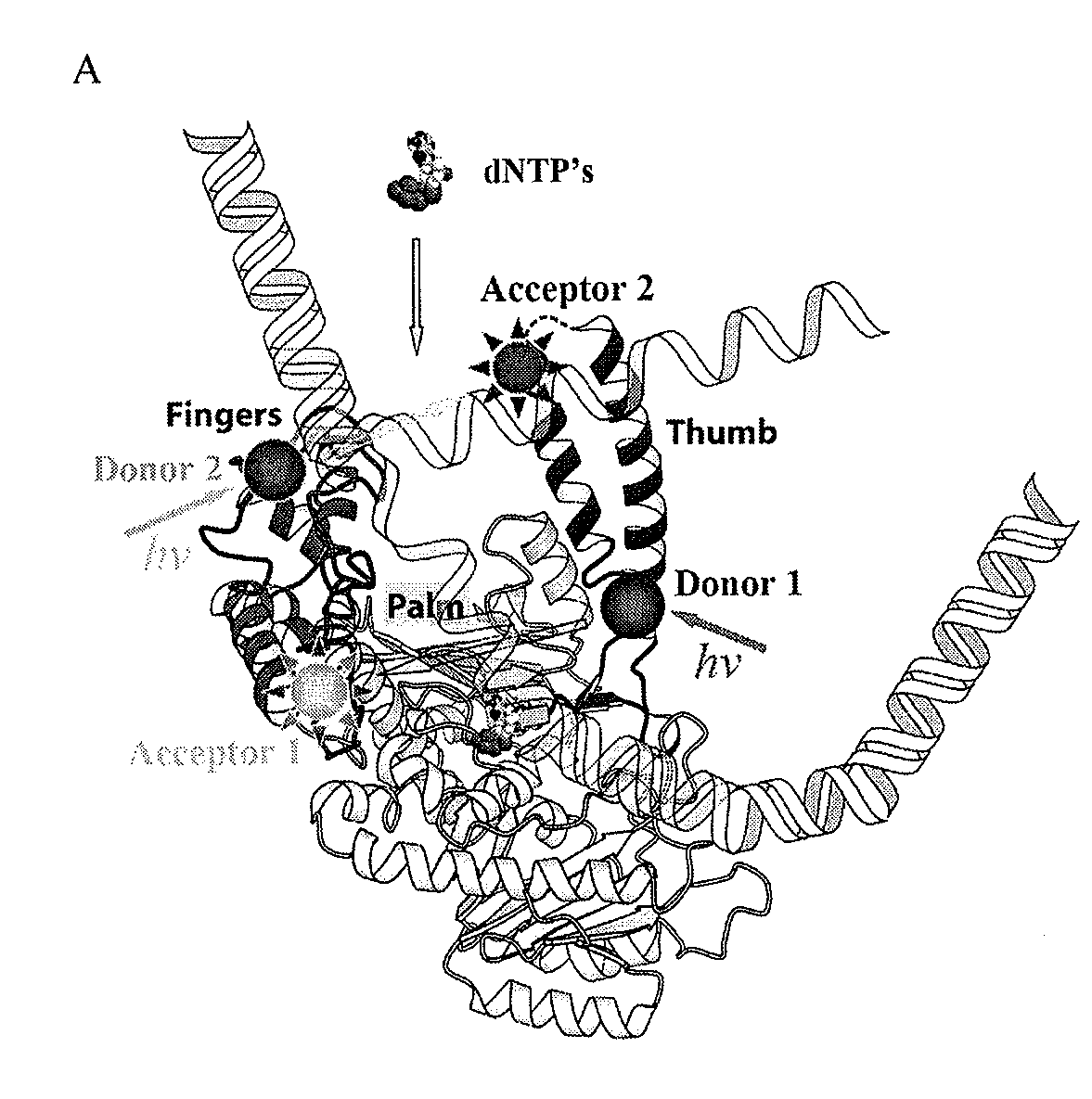
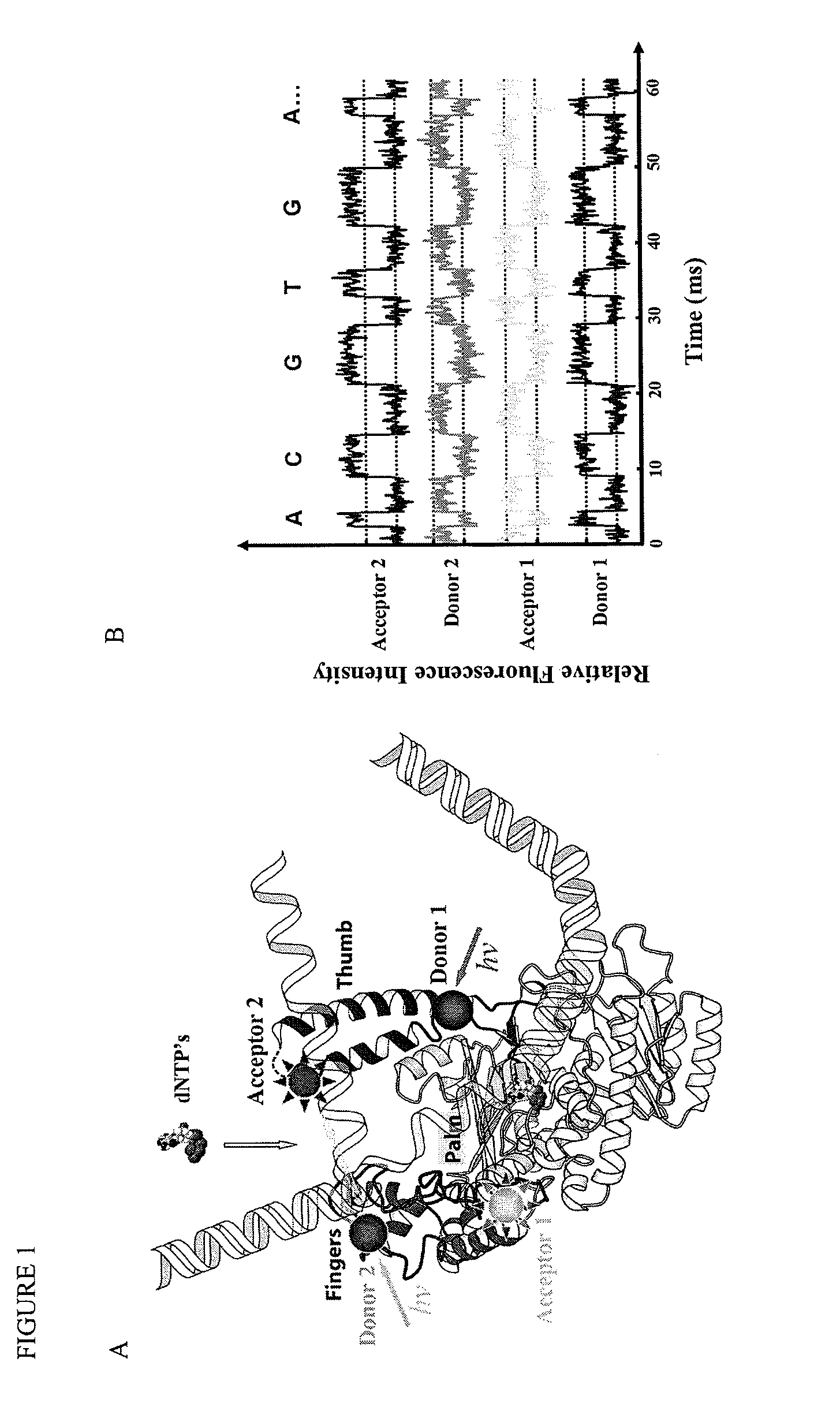
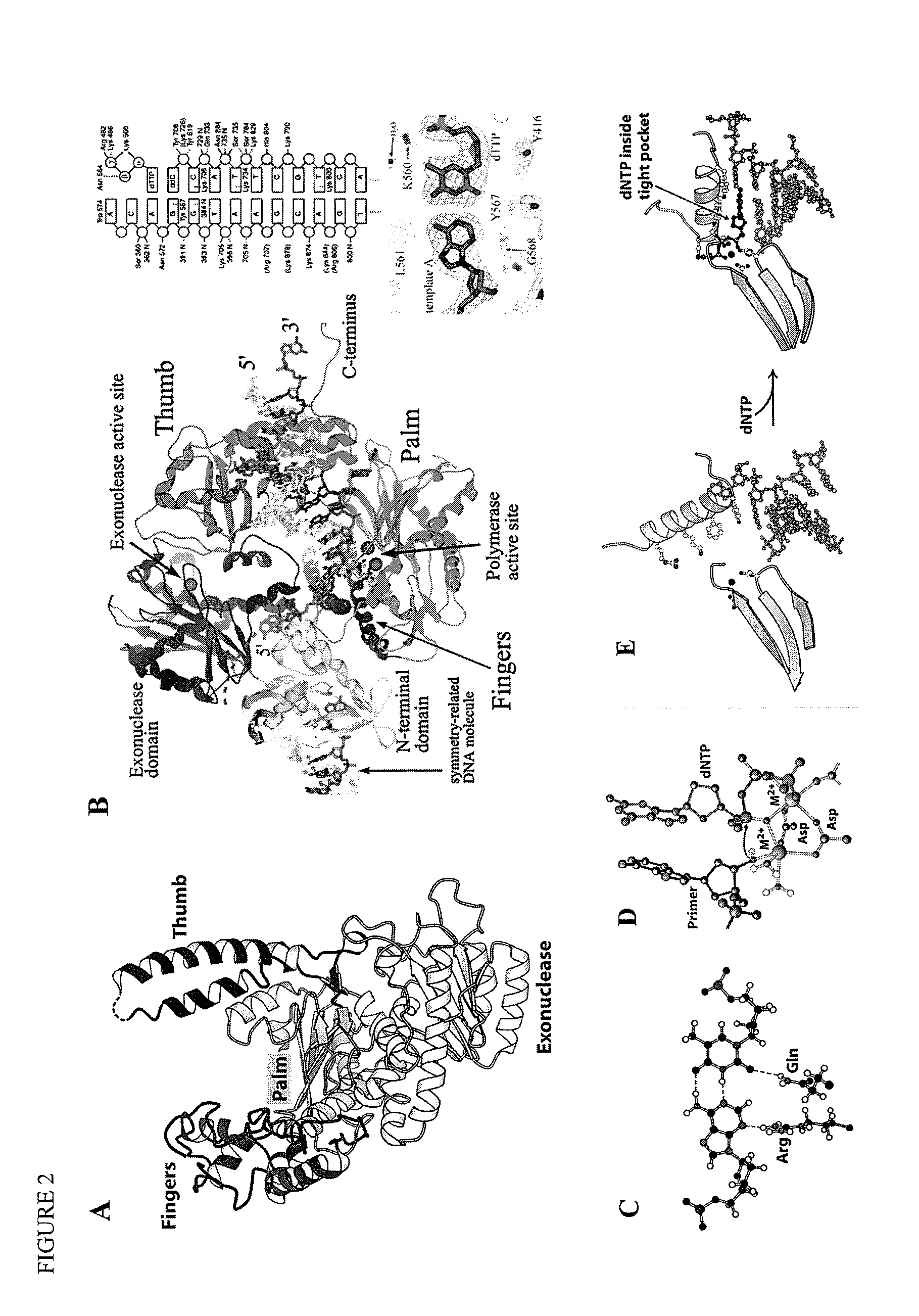
[0202]For the sake of illustration, we describe our systematic examination of the crystals structures of phi-29 DNA polymerase complexed with various substrates. We used a genetically engineered exonuclease-deficient phi-29 DNA polymerase (Berman et al., EMBO J, 26:3494-3505 (2007). The mutations involved in eliminating exonuclease activity do not affect the active site, or adjacent sites on the finger, thumb, and palm domains.
[0203]One of skill will appreciate that the sites disclosed for labeling phi-29 DNA polymerase can be applied to other DNA polymerases. As explained above, the structures of DNA polymerases are well-conserved. Thus, through optimal structural alignment (alignment of amino acids present in particular structural positions), the positions disclosed herein can be ascertained for a broad range of polymerases.
[0204]We selected residues on the finger subdomain as candidate labeling sites for the fluorescence donor and some on th...
example 2
In Vitro Translation of a Labeled DNA Polymerase
[0210]We have designed an efficient method for engineering labeled DNA polymerases with multiple labels positioned at designated residues. The general concept is illustrated in FIG. 6. For simplicity, the solid lines separating each named amino acid represent additional amino acids that are not shown.
[0211]The DNA polymerase coding sequence is cloned into a vector. The vector also includes regulatory sequences necessary for transcription (e.g. T7 promoter), translation initiation (ribosomal binding site—RBS, and start codon—ATG), and termination (stop codon—UAG). The codons encoding the targeted residues for labeling are mutated to a codon encoding a cysteine residue (TGC) using standard molecular biology methods.
[0212]The mRNA molecules are captured on a solid support by hybridization of a sequence at one end of the RNA molecules to a complementary oligonucleotide or PNA (peptide nucleic acid) immobilized on the solid support. The mRN...
example 3
READS Technology Using Immobilized DNA Polymerase
[0221]DNA polymerases are labeled as described above, and immobilized on glass coverslips. The surface of a glass coverslip is derivatized with a streptavidin. The glass coverslip is first cleaned with the RCA protocol, derivatized with amine group with aminoalkyl (e.g. gamma-aminopropyl) triethoxysilane, and then functionalized with biotin with NHS ester-PEG-biotin (e.g. NHS ester-PEG 5000-Biotin). The biotinylated surface can be patterned into highly-ordered arrays with feature size and spacing optimal for assembly of single molecular arrays and fluorescent imaging efficiency.
[0222]The biotinylated coverslip is then assembled into a flowcell for further functionalization with streptavidin. The biotinylated surface is functionalized with streptavidin by incubating the glass coverslip with a streptavidin solution, e.g. 1 μM streptavidin in a buffer solution such as phosphate buffer saline (PBS, 137 mM NaCl, 2.7 mM KCl, 10 mM sodium ph...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Förster radius | aaaaa | aaaaa |
| Förster radius | aaaaa | aaaaa |
| Förster radius | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com