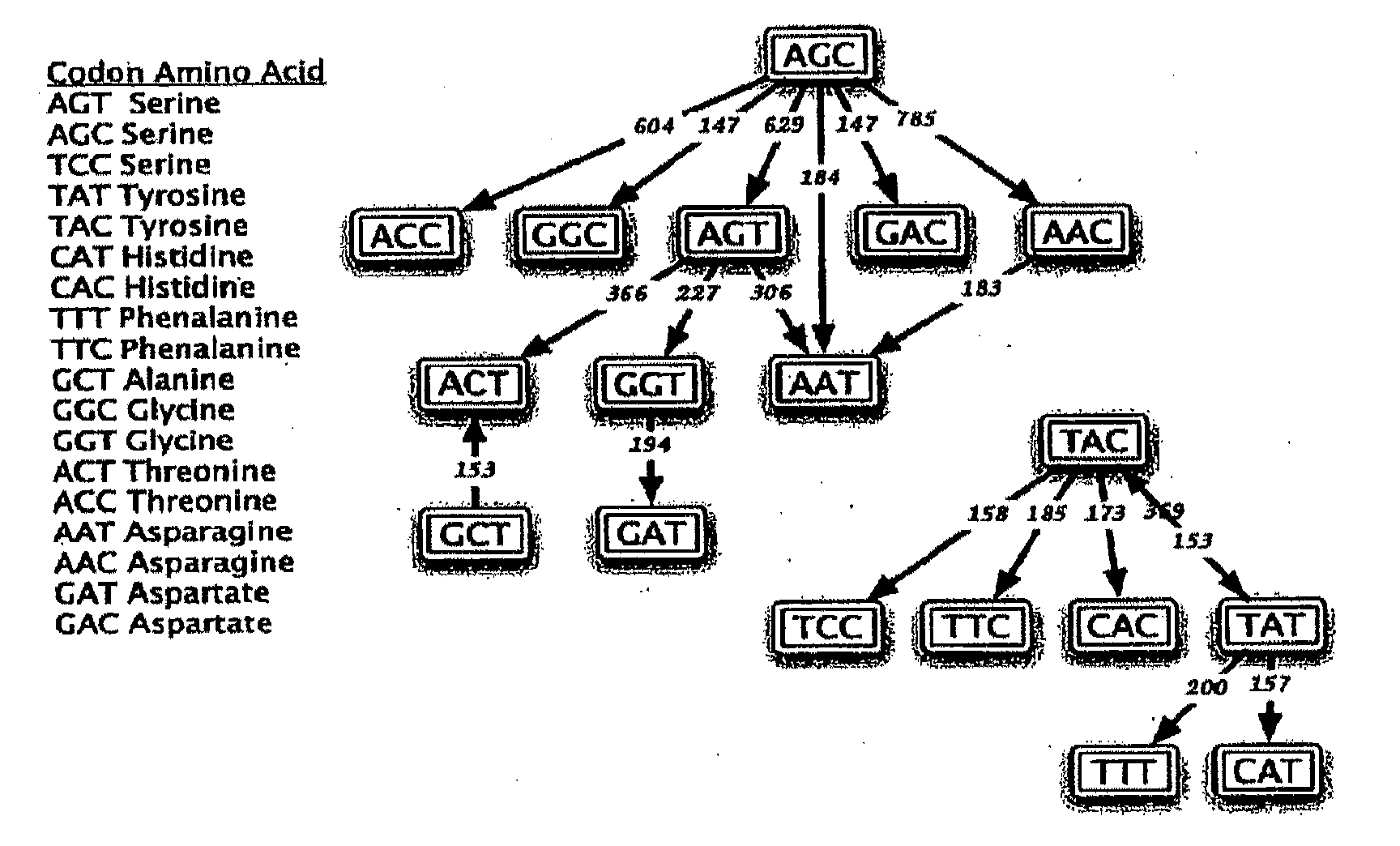
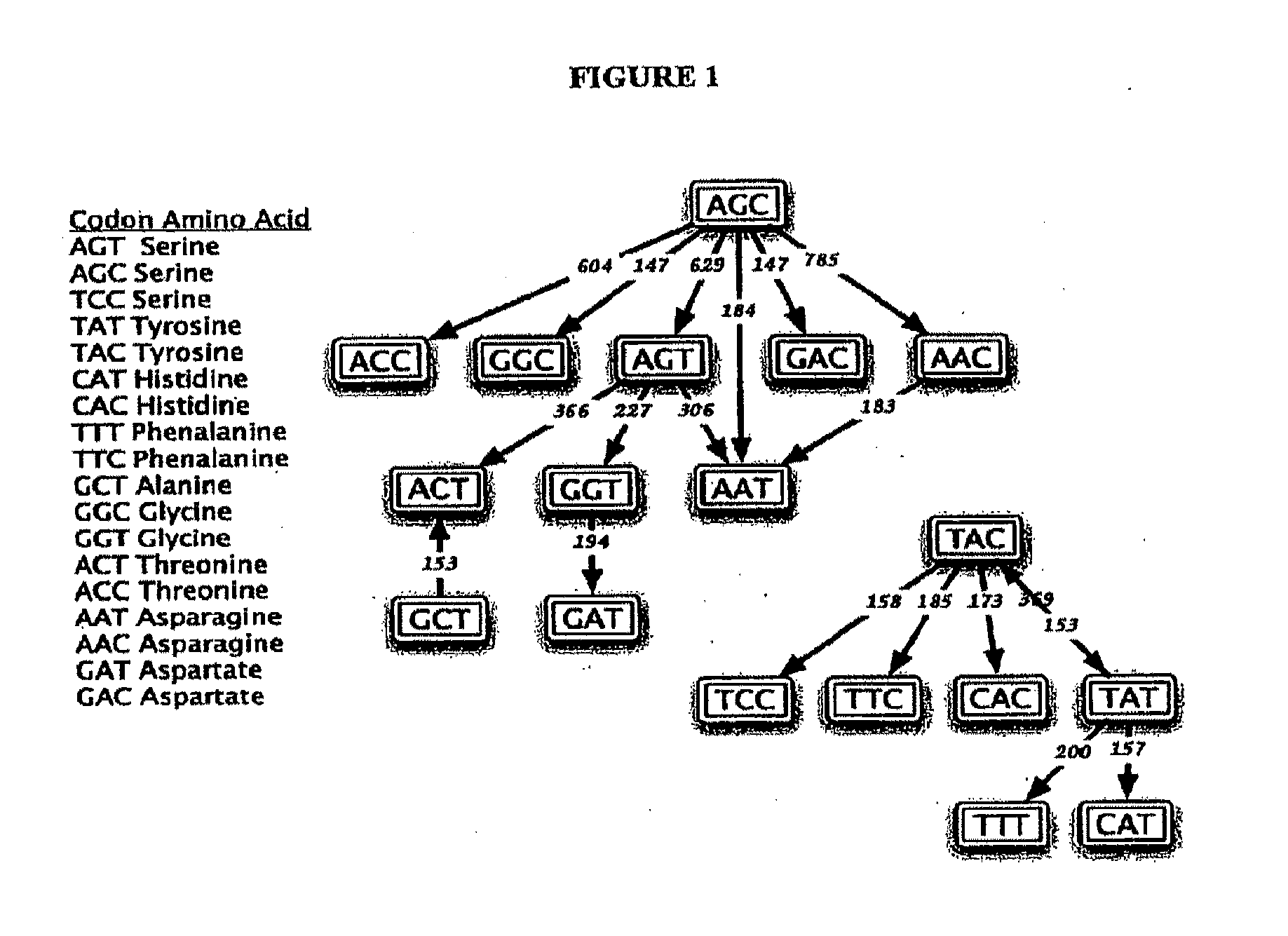
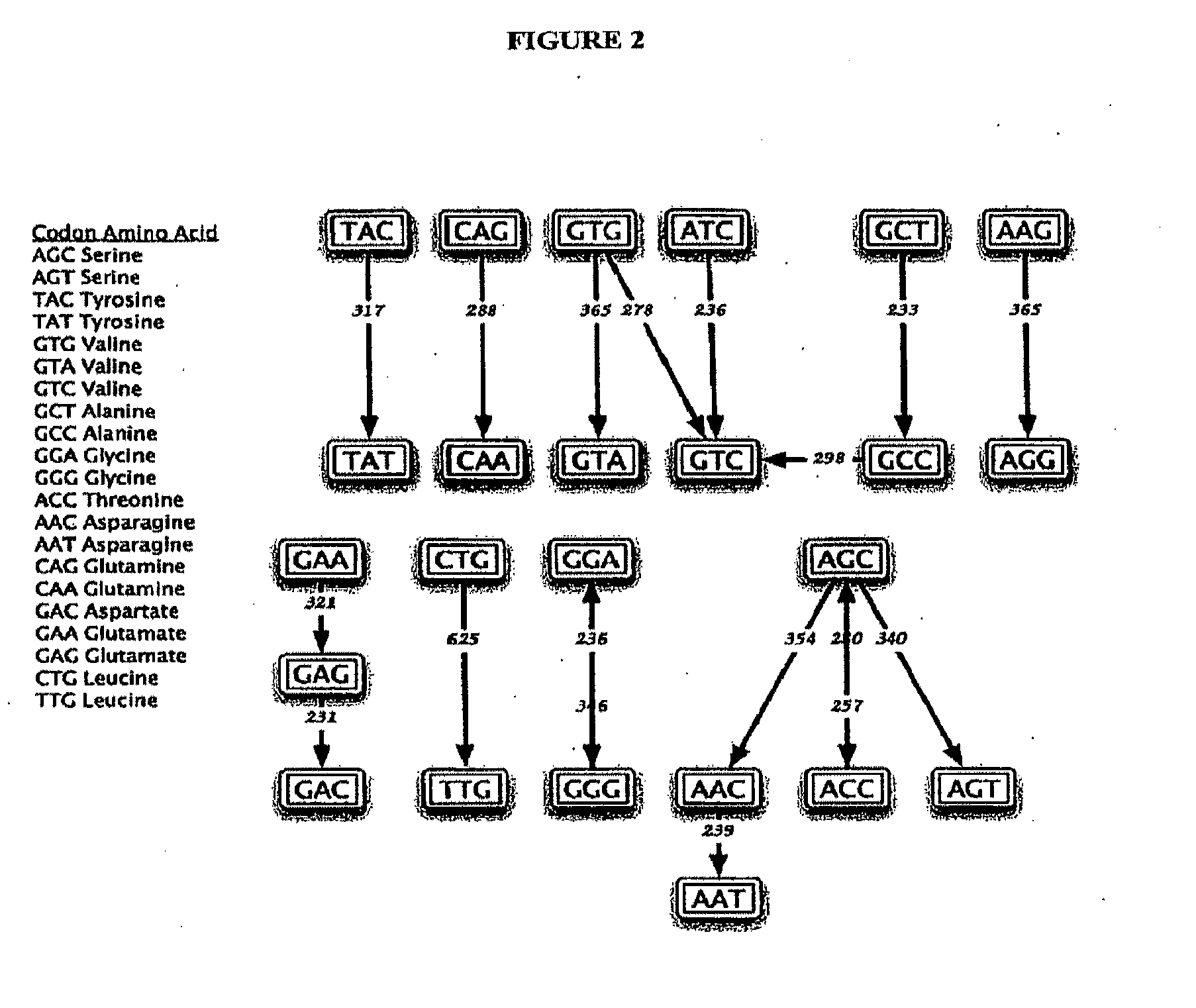
Somatic hypermutation systems
a somatic hypermutation and system technology, applied in the field of somatic hypermutation systems, can solve the problems of long development time, inefficiency, and system work against the more rapid development of improved therapeutics, and achieve the effects of preventing non-specific mutagenesis of structural proteins, stable maintenance of a mutagenesis system, and increasing and/or reducing shm
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Creation of Synthetic Polynucleotides Encoding Blasticidin
[0608]By decreasing the likelihood of somatic hypermutation in a vector element, such as a selectable marker, an enzyme involved in SHM, or a reporter gene, the vector and system for exerting and tracking SHM becomes more stable, thereby enabling somatic hypermutation to be more effectively targeted to a polynucleotide of interest.
[0609]A. Polynucleotide Design
[0610]In general, sequences are engineered for SHM using the teaching described herein, and as elaborated in sections III and IV. In the following examples, the sequence optimization is based on the hot spot and cold spot motifs listed in Table 7, and using the computer program SHMredesign.pl as described above.
[0611]Using this program, every position within the sequence is annotated with either a ‘+’, ‘−’, or ‘.’ symbol to designate whether it is desired to obtain a hotter, colder, or neutral changes in SHM susceptibility at that specific position. Where ‘+’ designates...
example 2
Creation of Synthetic Polynucleotides Encoding Hygromycin
[0630]A. Polynucleotide Design
[0631]The starting sequence for unmodified hygromycin is shown in FIG. 11, together with the initial analysis of hot spot and cold spot frequency.
[0632]As described for Example 1, sequence optimization is completed using the computer program SHMredesign, based on the hot spot and cold spot motifs listed in Table 7.
1. Cold Hygromycin
[0633]From iteration 1 to iteration 2000, an additional 71 cold spots are inserted into the gene, 12 existing hot spots are removed, and 61 CpG sites are removed making the gene sequence less susceptible to somatic hypermutation. No further beneficial changes are observed upon further iterations.
[0634]An optimized sequence for a SHM resistant version of hygromycin created using this approach is shown in FIG. 12, together with the resulting changes in frequency of hot spots and cold spots. Optimization of the hygromycin sequence to make the sequence more resistant to som...
example 3
Creation of Synthetic Polynucleotides Encoding Reporter Genes
[0642]A. Polynucleotide Design
[0643]The starting sequence for unmodified Teal Fluorescent Protein (TFP) is shown in FIG. 14, together with the initial analysis of hot spot and cold spot frequency.
1. Hot TFP
[0644]As described for Example 1, sequence optimization is completed using the computer program SHMredesign, based on the hot spot and cold spot motifs listed in Table 7; the resulting hot and cold versions of TFP are shown in FIGS. 15 and 16, respectively.
[0645]Optimization of the TFP sequence to make the sequence more susceptible to somatic hypermutation resulted in an increase of about 170% in number of hot spots (an increase of 28), and reduced the number of cold spots by about 26% (a decrease of 27). Overall the frequency of hot spots increased to an average density of about 10 hot spots per 100 nucleotides from an initial density of about 6 hot spots per 100 nucleotides, and the overall frequency of cold spots decr...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
| Fraction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com