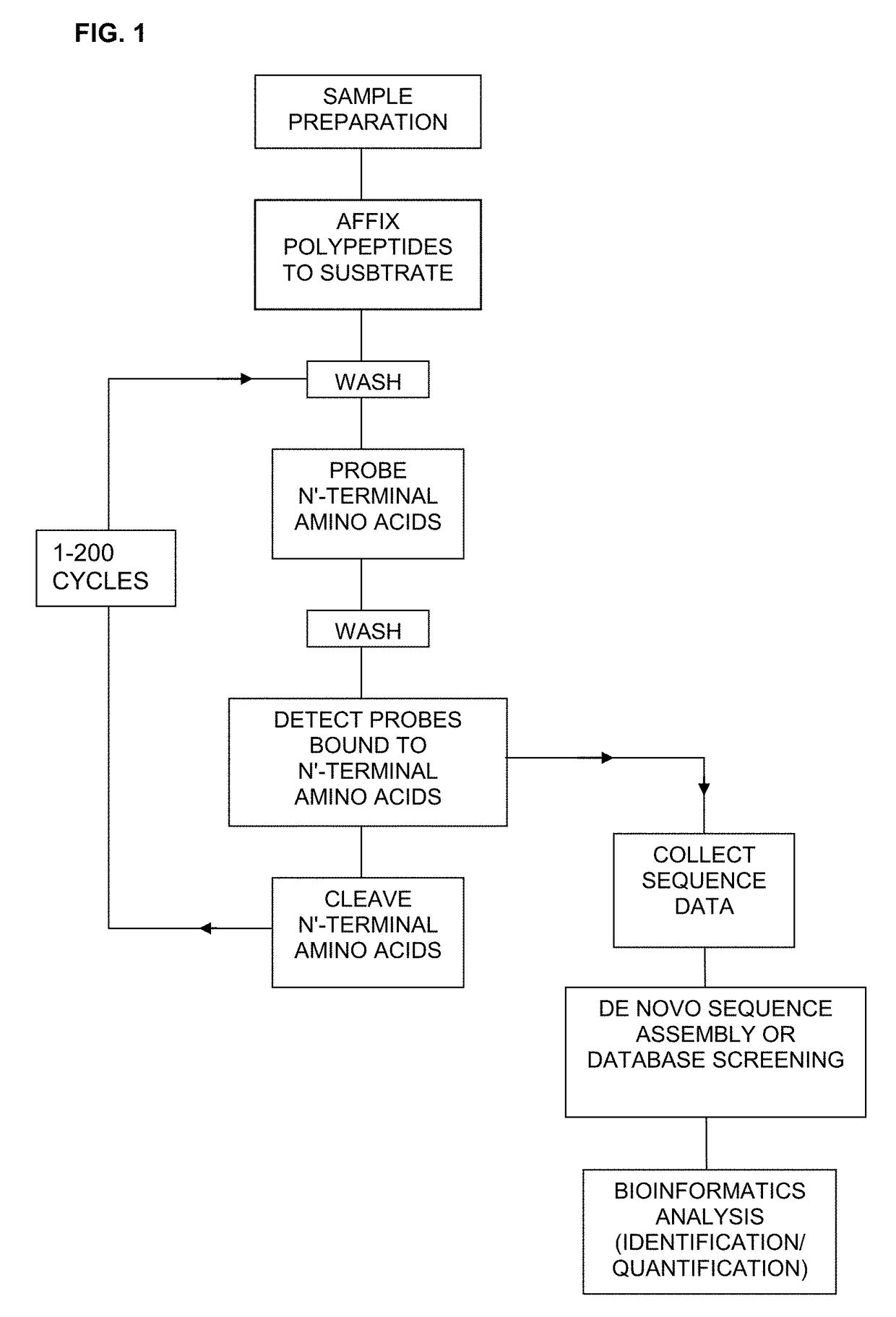
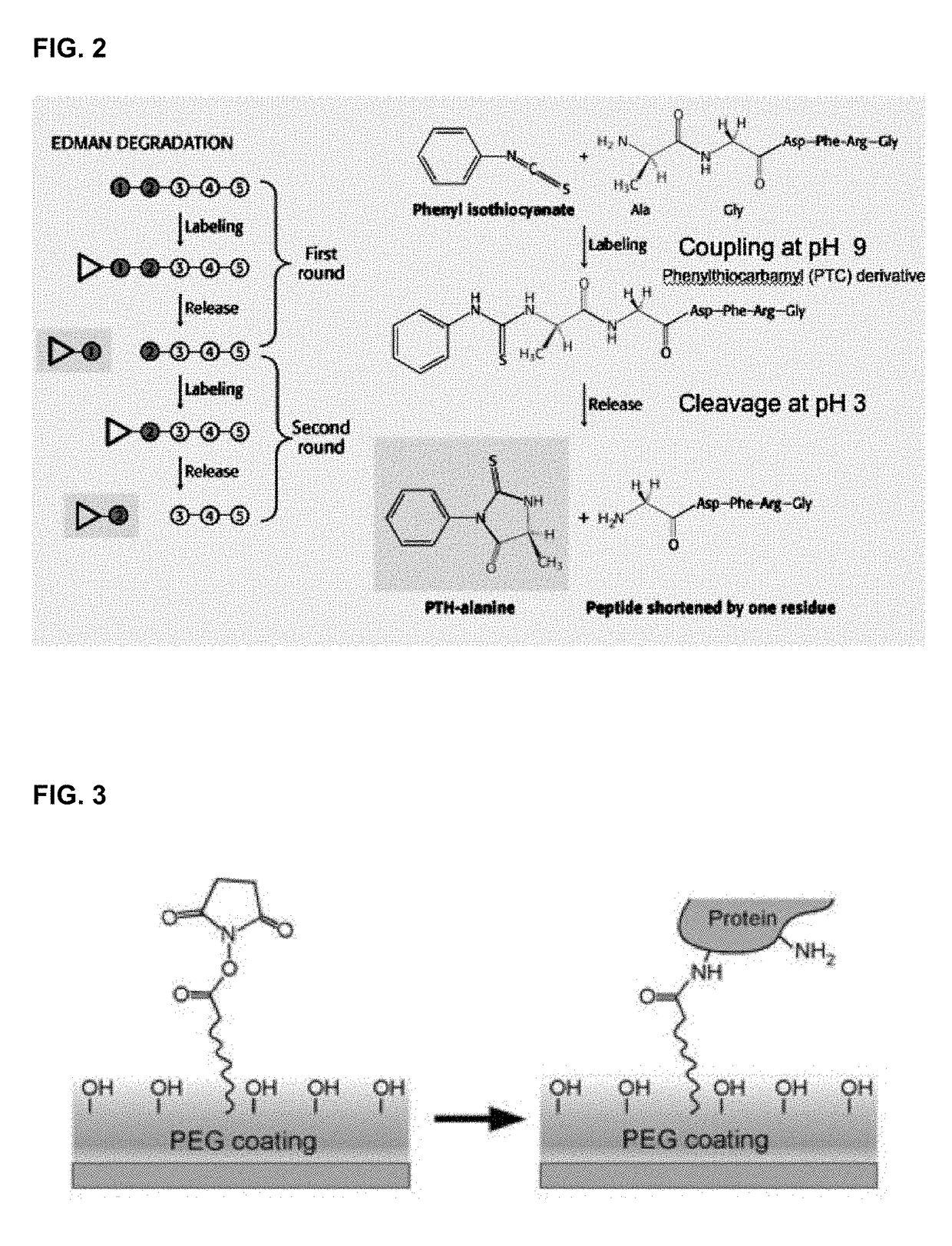
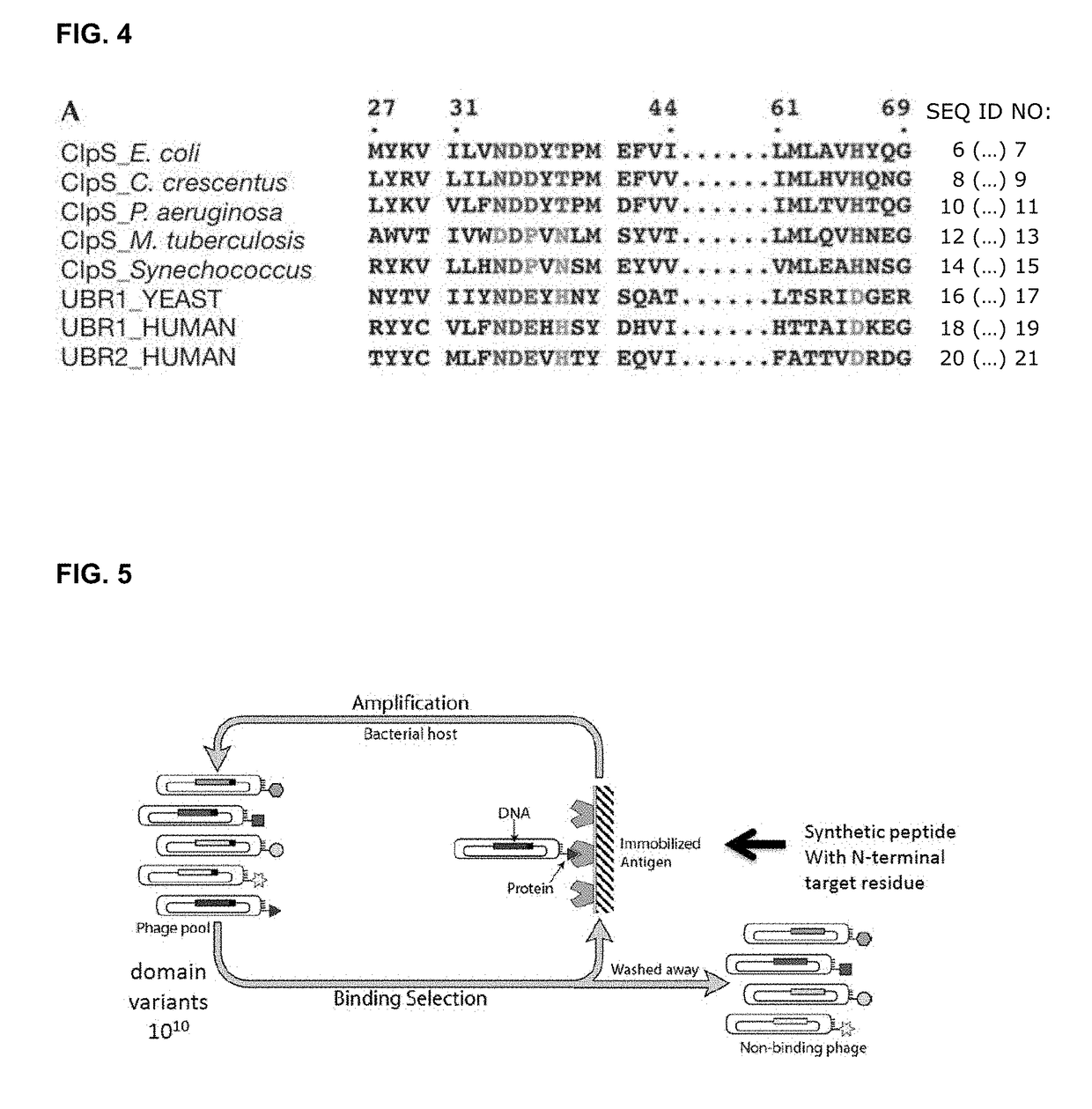
Protein sequencing method and reagents
a technology of protein sequencing and reagents, applied in the field of protein sequencing, can solve the problems of large sample amounts, low method throughput, computational complexity,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Affinity Capture Reagents Based on the CIpS Adaptor Protein Scaffold
[0132]Phage display and combinatorial site-directed mutagenesis is used to identify variants of the natural N-terminal amino acid binding pocket or structural domain of the CIpS adaptor protein family that selectively bind N-terminal amino acids. Structural modeling of the binding pocket and protein engineering are used to further define modified variants of CIpS family members that are suitable for use with the methods described herein.
Results
[0133]Modified CIpS adaptor proteins are selected using screening procedures familiar to those trained in the art. Affinity capture reagents are subsequently identified that exhibit high affinity / selectivity by phage display to N-terminal amino acids.
example 2
Single Molecule Sequencing of a Synthetic Polypeptide
[0134]An artificial test polypeptide comprising of the heptapeptide amino acid sequence TyrPheArgTyrPheArgLys (SEQ ID NO: 5) is synthesized. The polypeptide is affixed to a substrate via its C-terminal amino acid carboxy group or the lysine side chain. The substrate is then washed in order to remove any debris. Probes containing an N-terminal affinity capture reagent identified as shown in Example 1 are coupled to a fluorescent moiety. The probes are then added to the substrate under conditions that encourage the binding of the probes to polypeptide. The substrate is then washed to remove any non-specifically bound probes. The probe bound to a single affixed polypeptide is then detected using an optical detection system. The identity of the probe bound to the polypeptide is recorded and the N-terminal amino acid of the polypeptide is cleaved via Edman degradation. Additional rounds of probes are then added to the substrate in orde...
example 3
Validation of Assay Conditions and Protocols
[0136]Variants of CIpS are derived via structural modeling, docking, combinatorial site-directed mutagenesis, and / or the experimental selection of high affinity and high specificity binders by phage display as shown in Example 1.
[0137]Recombinant CIpS protein is prepared for use as a probe by expression in E. coli, or other expression system, and purified using standard biochemical methods, and subsequently coupled with one or more quantum dots with defined absorbance and emission wavelengths, including near infrared fluorescence emitters. The labels can be coupled to an N-terminal region of CIpS that is distinct from the C-terminal domain that serves as the actual peptide ligand (i.e. N-terminal amino acid) binding pocket.
[0138]A glass or polystyrene substrate is coated with PEG / NHS, or equivalent reactive carbohydrate linker, to minimize non-specific adsorption and spurious background signal.
[0139]The test proteins and peptides include a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com