Methods for detection of nucleic acid sequences in urine
a nucleic acid sequence and urine technology, applied in the field of non-invasive methods, can solve the problems of ineffective methods, inconvenient diagnosis of certain genetic defects, and intrusion of genetic material for diagnosis in certain cases, and achieve the effect of reducing dna degradation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
DETECTION OF POLYMERIC DNA IN URINE OF MICE PREINOCULATED WITH λ PHAGE DNA
[0161]This example analyzes the ability of DNA to cross the kidney barrier in rodents and appear in detectable form in urine.
[0162]λ phage DNA (New England BioLabs, MA) was labeled by nick translation with [α-32P] dNTP DNA (New England BioLabs, MA) using the Klenow fragment of E. Coli DNA polymerase to a specific radioactivity of 108 cpm / μg as previously described. ( / ]Sambrook J., Fritsch E. F., Maniatis T., Molecular Cloning. A Laboratory Manual. 2d Edition. Cold Spring Harbor Laboratory Press, 1989). Two months old male Wistar rats were injected subcutaneously with 0.4 μg of the [32p] labeled DNA. Urine samples were then collected for three days and total and acid-insoluble radioactivity was measured in a liquid scintillation counter. The kinetics of excretion of acid-insoluble radioactivity in urine appear in Table 1, below. It was determined that 3.2% of the total DNA with which the rats were inoculated cr...
example 2
DETECTION OF HUMAN DNA SEQUENCES IN URINE OF MOUSE PREINOCULATED WITH HUMAN CELLS
[0167]Example 1 showed that DNA sequences could remain in polymeric form in the blood stream, cross the kidney barrier, and remain suitable for subsequent detection. The next set of experiments were performed to determine if DNA from cells dying in the organism but not in the urinary tract can be detected in urine.
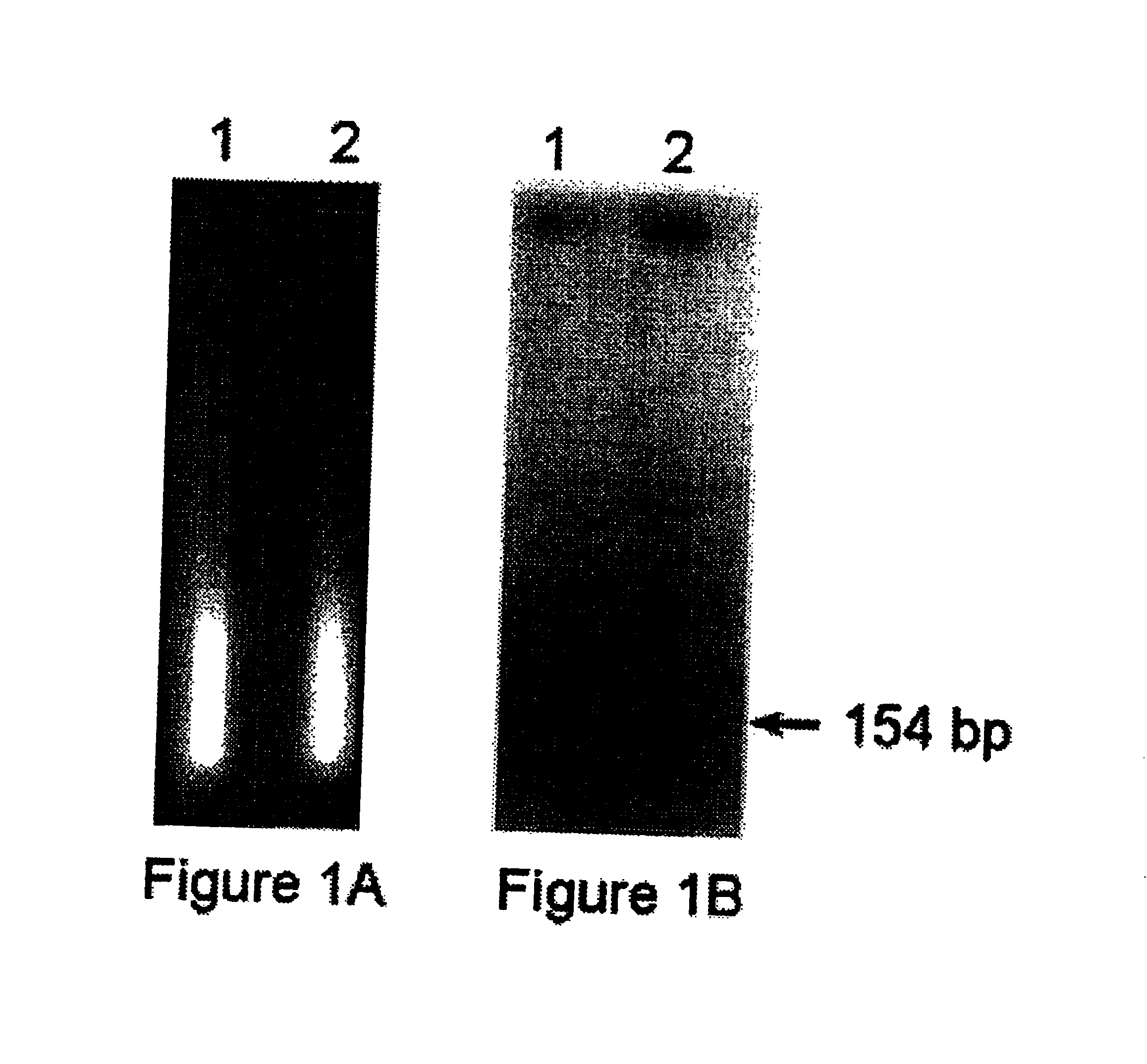
[0168]Human Raji lymphoma cells growing in RPMI supplemented with 5% fetal calf serum were irradiated with 1000 rads of 137CS γ-rays. Mice were then inoculated subcutaneously with 108 cells each. Urine samples were collected for three days, and DNA was isolated as described above. Human-specific DNA sequences were detected by multilocus screening using Alu oligonucleotide-directed PCR as previously described, (Zietkiewicz, E., Labuda M., Sinnett D., Glorieux F. H., and Labuda D., Proc. Natl. Acad. Sci. USA, 89, 8448-8451, 1992) followed by electrophoresis in a 1.5% agarose gel.
[0169]The result...
example 3
DETECTION OF TRANSRENAL NUCLEIC ACIDS
[0171]Taken together, Examples 1 and 2 demonstrate that, in the mouse model, both free DNA and DNA from dying cells cross the kidney barrier and can be detected in urine by PCR analysis. Two systems were selected as models to demonstrate that DNA can cross the kidney barrier and remain in polymeric form in human urine samples. The systems are designed to focus on DNA originating from cells dying outside the urinary tract rather than DNA that appears in urine due to the death of cells in the urinary tract.
[0172]Women who were either pregnant or transfused with male blood were studied because both of these models represented humans having DNA in their bodies that is not present in their normal genome. Each woman studied was analyzed for the presence of Y chromosome-specific sequences in their urine.
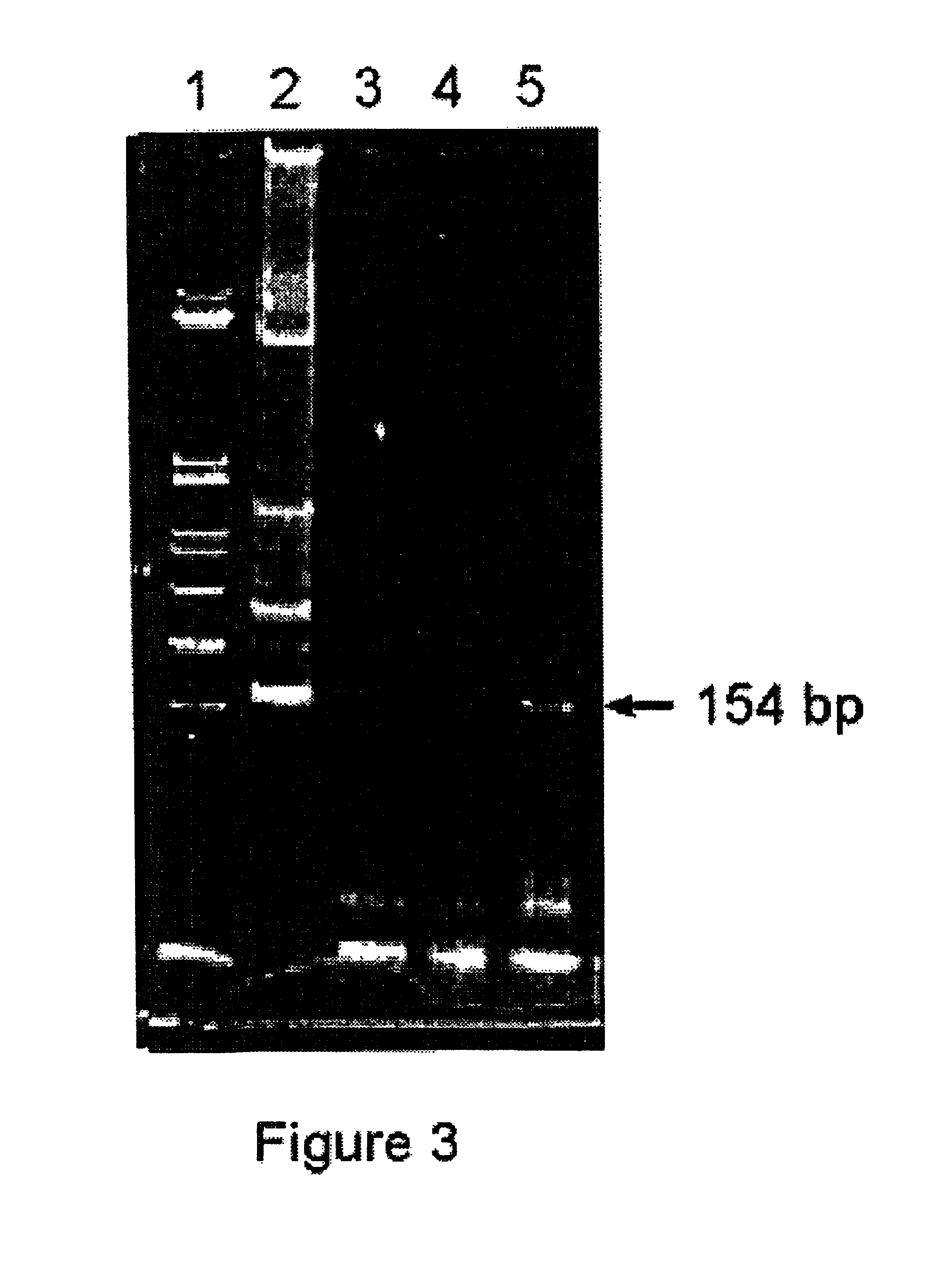
Detection of repeated and Single Copy Y Chromosome Specific Sequences in the Urine of Women Pregnant with Male Fetuses.
[0173]As discussed above, apoptot...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| Tm | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com