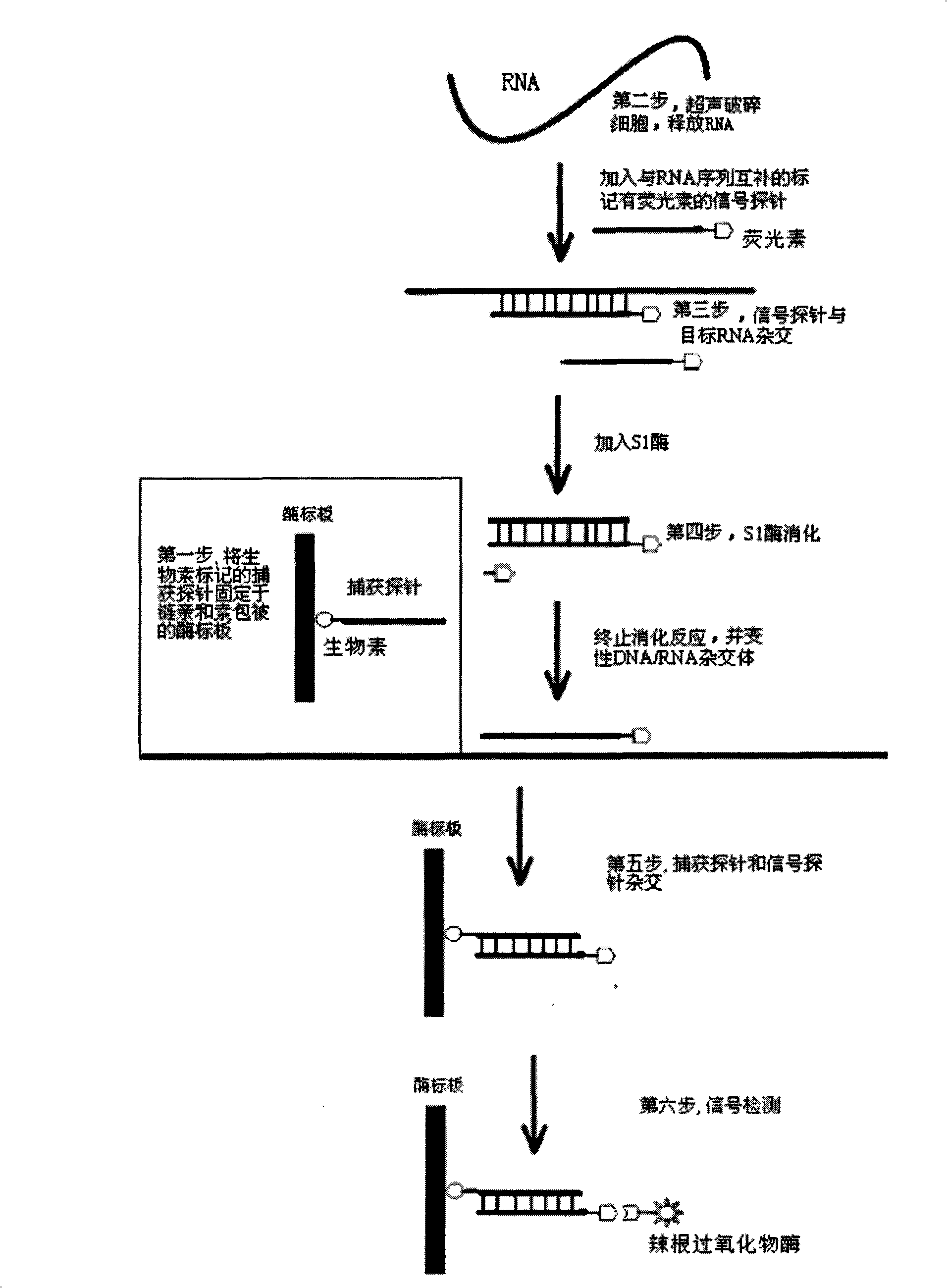
RNA quantitative determination method utilizing S1 enzyme cutting single-chain nucleic acid characteristic
A quantitative detection method and cutting sheet technology, applied in the field of bioengineering, can solve the problems of cumbersome steps in sandwich hybridization technology, lack of specificity of detection, increase of detection cost, etc., and achieve stable results, simple method, and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
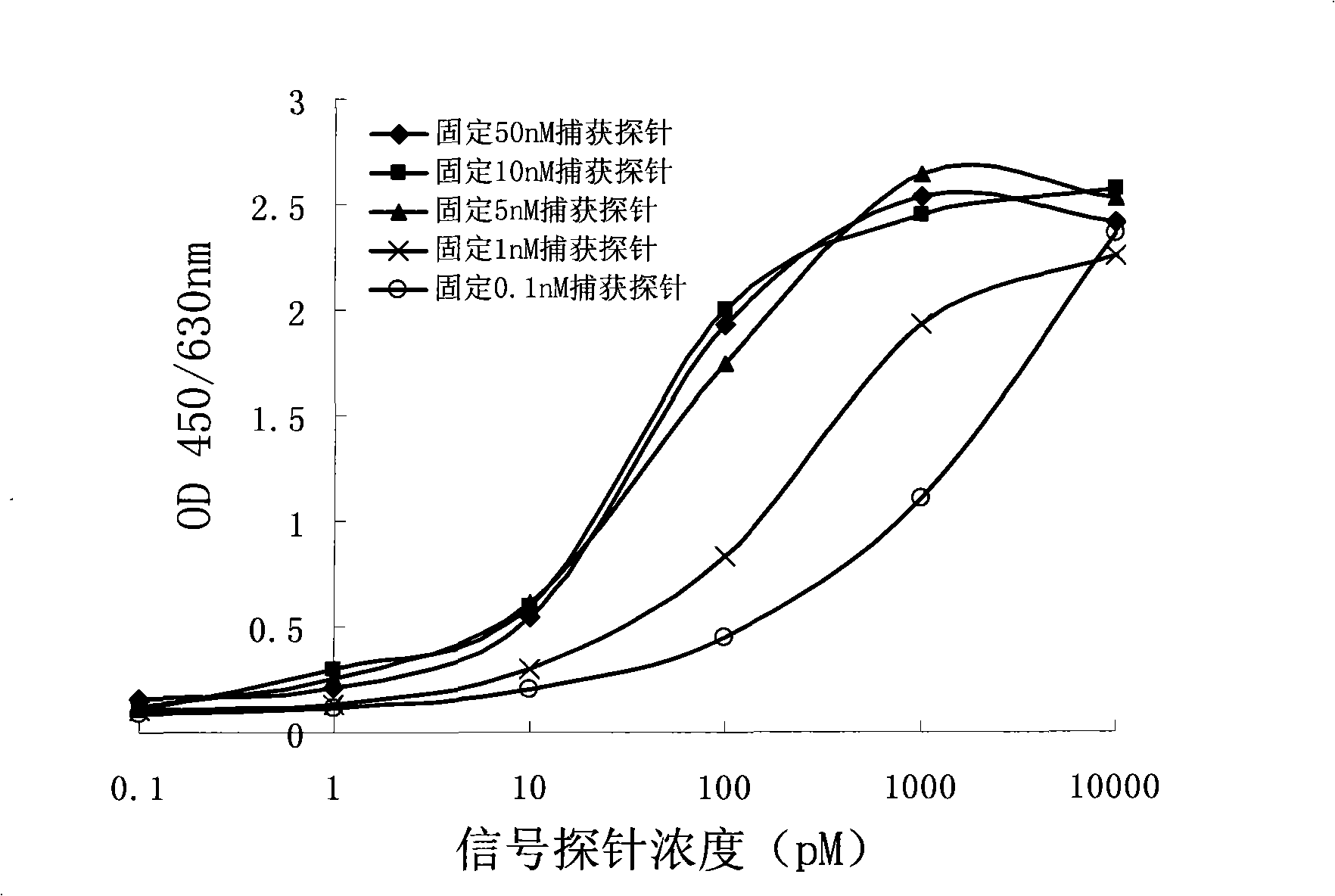
[0028] Effects of different concentrations of capture probes on the detection signal:
[0029] 1.1. Design signal probes and capture probes for Pseudomonas fluorescens:
[0030] Search the sequence of AY394845 in NCBI, and get AY394845, Pseudomonas sp.M18 16Sribosomal RNA gene, partial sequence. Input the obtained sequence into the software primerprimier 5 for probe search, set the probe length to 40+-3bp (other parameters default to ), select probes with GC content of 40-60% according to rating from high to low in the obtained probes, and obtain 39 bases of probes against M1816s RNA:
[0031] Capture probe:
[0032] 5'-(biotin)-AGGGTGCAAGCGTTAATCGGAATTACTGGGCGTAAAGCG
[0033] Signal probe:
[0034] 5’-(FAM)-CGCTTTACGCCCAGTAATTCCGATTAACGCTTGCACCCT
[0035] 1.2. Capture probe immobilization:
[0036] Dilute with phosphate buffer solution to obtain capture probes with a concentration of 0.1nM, 1nM, 5nM, 10nM and 50nM, respectively take 100μl and add them to the enzyme plate...
Embodiment 2
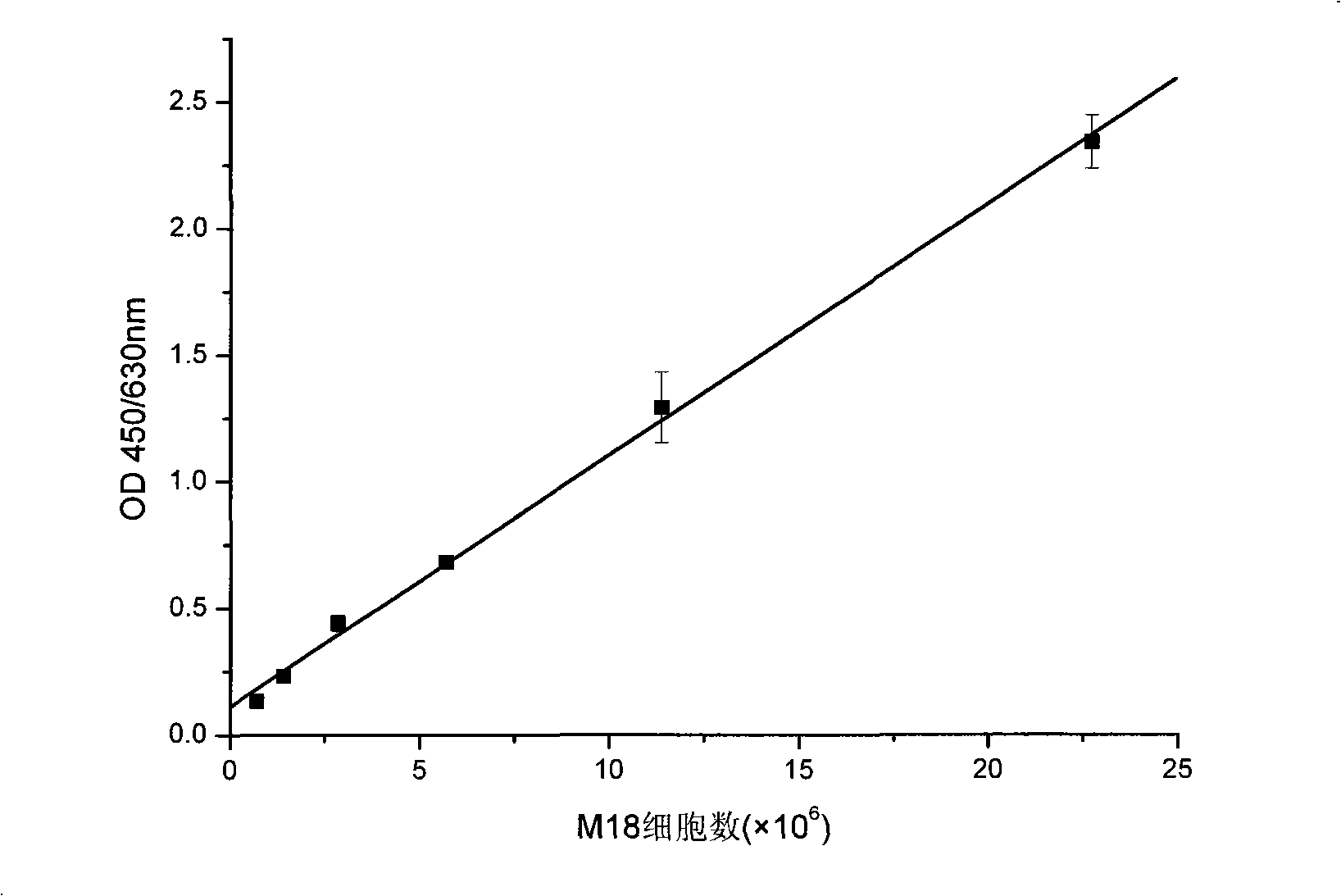
[0046] The detection limit and linear range of the 16s RNA of Pseudomonas fluorescens M18 (the Pseudomonas fluorescens strain M18 used has been designated by the Chinese Patent Office as a depository unit: Beijing, China Microorganism Culture Collection Management Committee General Microbiology Center, deposit number CGMCC NO.0462, 2000.6.27):
[0047] 2.1. Design signal probes and capture probes for Pseudomonas fluorescens:
[0048] Search the sequence of AY394845 in NCBI, and get AY394845, Pseudomonas sp.M18 16Sribosomal RNA gene, partial sequence. Input the obtained sequence into the software primerprimier 5 for probe search, set the probe length to 40+-3bp (other parameters default to ), select probes with GC content of 40-60% according to rating from high to low in the obtained probes, and obtain 39 bases of probes against M18 16s RNA:
[0049] Capture probe:
[0050] 5'-(biotin)-AGGGTGCAAGCGTTAATCGGAATTACTGGGCGTAAAGCG
[0051] Signal probe:
[0052] 5'-(FAM)-CGCTTTAC...
Embodiment 3
[0070] Determination of 16s RNA content in different growth stages of Pseudomonas fluorescens M18:
[0071] 3.1. Bacterial culture and plate count:
[0072] Inoculate the plate-activated M18 strain into 100ml of KB liquid medium, culture it on a shaker at 28°C and 160rpm, and use the plate counting method to detect the bacterial concentration at 12 hours, 24 hours, 36 hours and 60 hours, and liquid nitrogen quick-freezing Save the bacteria. The bacterial concentration measured by the plate counting method was 0.97×10 10 CFU / ml, 2.5×10 10 CFU / ml, 7.4×10 10 CFU / ml and 9.5×10 10 CFU / ml.
[0073] 3.2. Capture probe immobilization:
[0074] Add 100 μl of 5 nM capture probe diluted with phosphate buffer to a streptavidin-coated microtiter plate, bind at 37°C and 200 rpm for 2 hours, wash with phosphate buffer 3 times, and store at 4°C for use.
[0075] 3.3. Bacterial fragmentation:
[0076] Take a portion of the M18 cells (centrifuged from 1ml of the bacteria liquid) that we...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com