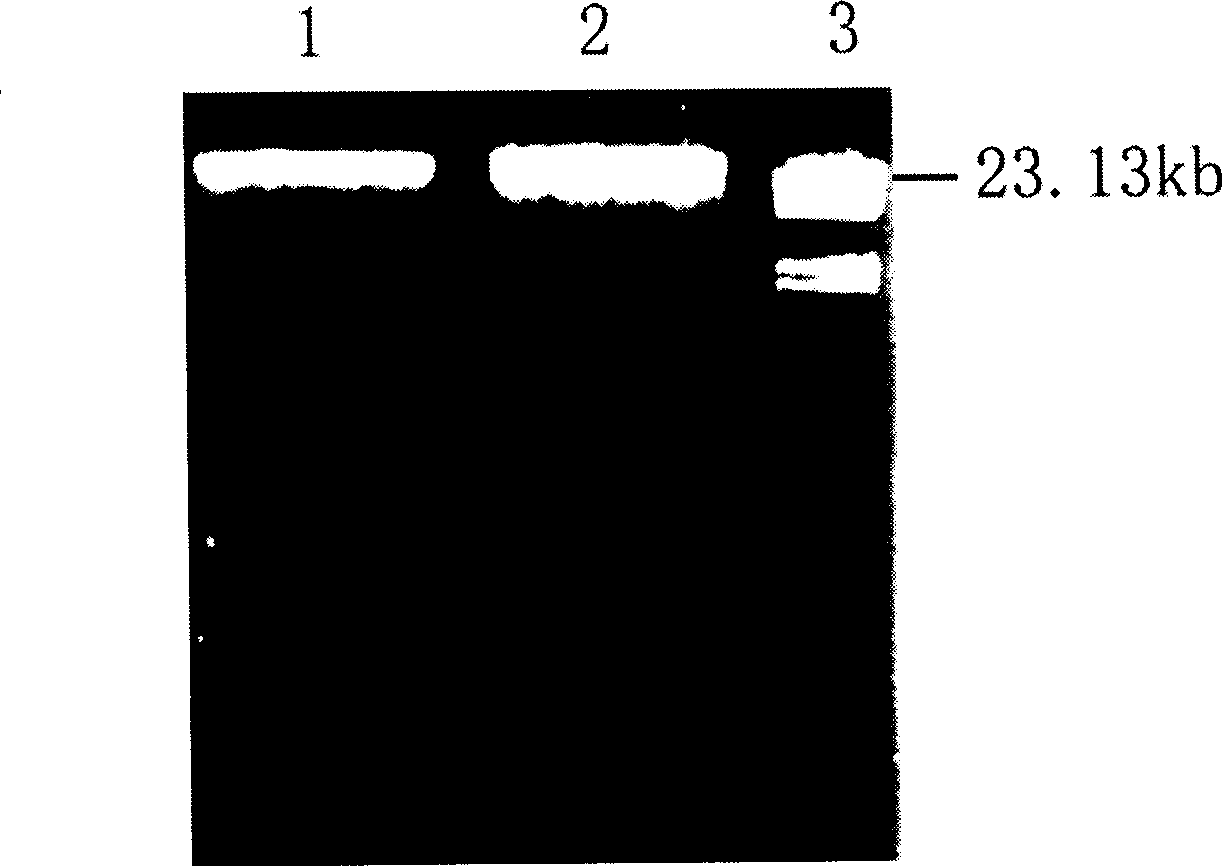
Gene encoding beta-glucosidase
A technology of glucosidase and gene, applied in the field of enzyme gene, can solve the problems of cloning β-glucosidase gene and less microbial research
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0064] Example 1. A large amount of metagenomic DNA was directly extracted from soil samples.
[0065] 1) Weigh 4-4.5g of alkaline soil, grind it finely, add 10mL of 2 parts of TENS (each part is: 100mM Tris-HCL, 40mM EDTA, 200mMNaCI, 2% SDS, pH 8.0), in a shaker mix thoroughly;
[0066] 2) Add 100 μL of 25 mg / mL Protease-K (Protease-K), dissolve in a 70°C water bath for 30 minutes, and stir and mix once every 10 minutes;
[0067] 3) Then place it in liquid nitrogen for quick freezing for 2.5 minutes, quickly place it in a low-temperature refrigerator at -20°C for 5 minutes, and immediately dissolve it completely in a boiling water bath after taking it out. Step 3) Repeat 3 times in total;
[0068] 4) Put it still at room temperature for 3 minutes, then centrifuge, collect the supernatant and put it on ice, add 10mL of Buffer A (composition: 50mM Tris-HCL, 25mM EDTA, 3% SDS, 1.2% After PVPP, pH 8.0), centrifuge to collect the supernatant; mix all the supernatants together a...
Embodiment 2
[0073] Example 2. Purification of crude metagenomic DNA by electroelution.
[0074] This method can recover DNA fragments of any size, especially DNA fragments larger than 5kb. Moreover, the success rate is extremely high, and it is easy to purify.
[0075] (1) Treatment of dialysis bags
[0076] A. Cut out a dialysis bag with a length of 10 to 20 cm;
[0077] B, the dialysis bag was boiled in 2% (w / v) (mass / volume) sodium bicarbonate, 1mM EDTA (pH8.0) solution for 10 minutes;
[0078] C. Take out the dialysis bag and rinse thoroughly with distilled water;
[0079] D. Boil in 1mM EDTA solution (pH 8.0) for 10min (do not pour out the solution);
[0080] E. Allow the dialysis bag to cool naturally and store at 4°C (note that during storage, the dialysis bag should always be immersed in the solution to prevent drying;
[0081] F. Before use, fully rinse the inner and outer walls of the bag with distilled water;
[0082] Note: Wear medical latex gloves to operate.
[0083] ...
Embodiment 3
[0093] Embodiment 3, the construction of the genomic library of uncultivated microorganisms in alkaline polluted soil
[0094] Firstly, the modified alkaline denaturation method was used to extract a large amount of plasmid DNA from the cloning vector pGEM-3Zf(+) to obtain relatively high-purity plasmid DNA. Its main form is supercoiled (CCC), which was digested by EcoR I enzyme and turned into a linear DNA molecule. , the linear pGEM-3Zf(+) plasmid DNA was treated with CIP dephosphorylase, and then directly used to connect partially digested soil metagenomic DNA.
[0095] The purified metagenomic DNA was partially digested with EcoR I, the digested products were separated by agarose electrophoresis, these fragments were ligated with pGEM-3Zf(+) DNA treated with CIP dephosphorylase, and the ligated products were transformed by electrophoresis Methods Introduce Escherichia coli DH5 α in the presence of X-gal (5-bromo-4-chloro-3-indole-β-D-galactoside) (40 μg / mL), IPTG (isopropy...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com