Method for quantitative detection of mycoplasma hyopneumoniae
A technology of Mycoplasma hyopneumoniae and a detection method, applied in the field of real-time fluorescent quantitative PCR primers and probes, can solve the problems of difficult Mhp accurate quantitative detection and the like
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] The establishment of embodiment 1Mhp TaqMan fluorescent quantitative PCR detection method
[0028] This example aims to use primers to p110realF / R and probes for quantitative PCR, and establishes a Mhp TaqMan fluorescent quantitative PCR detection method.
[0029] 1.1 Design and synthesis of primers and probes
[0030] According to the p110 (p76) gene sequence of Mycoplasma hyopneumoniae genome published on GenBank (ACCESSION No.NC_006360), the following fluorescent quantitative PCR primers and probes were designed using Primer Express v2.0 software.
[0031] P110realF: AGGATACAAACTGAGAAACCGAGCTA
[0032] P110realR: CAAGACCGAGTGGGTATGACCT
[0033] Probe: TGGACAGATCGGTGATACAACCCCACA
[0034] 1.2 Extraction of Mycoplasma hyopneumoniae genome
[0035] 1) Centrifuge 1ml Mhp Mycoplasma culture at 12000rpm room temperature for 10min, discard the supernatant;
[0036] 2) Add 300 μl proteinase K lysis buffer system (10 mM Tris-EDTA [pH=7.5], 0.5% [wt / vol] SDS, 100 μg / ml pr...
Embodiment 2
[0098] The specificity test of embodiment 2 Mycoplasma hyopneumoniae fluorescent PCR detection method
[0099] Common genetically engineered bacteria (Escherichia coli DH5α strain and BL21 strain), oral mycoplasma, and pathogens often mixed with Mycoplasma hyopneumoniae (porcine circovirus, PRRSV and mycoplasma hyorhina) were used as control samples, respectively. The standard plasmid was used as a positive control, and the sterilized DDW was used as a negative control to test the specificity of the fluorescent PCR detection method based on Mycoplasma hyopneumoniae.
[0100] 2.1 Experimental materials
[0101] For comparison, Mycoplasma orale CVCC 379 strains and Mycoplasma hyorhinis CVCC 361 strains were purchased from China Veterinary Drug Control Institute, and the others were laboratory-preserved strains.
[0102] 2.2 Sample pretreatment
[0103] 2.2.1 Acquisition of viral DNA
[0104] PRRSV RNA was extracted according to the instructions of Invitrogen Company. Oligo(d...
Embodiment 3
[0127] The sensitivity test of embodiment 3 Mycoplasma hyopneumoniae fluorescent PCR detection method
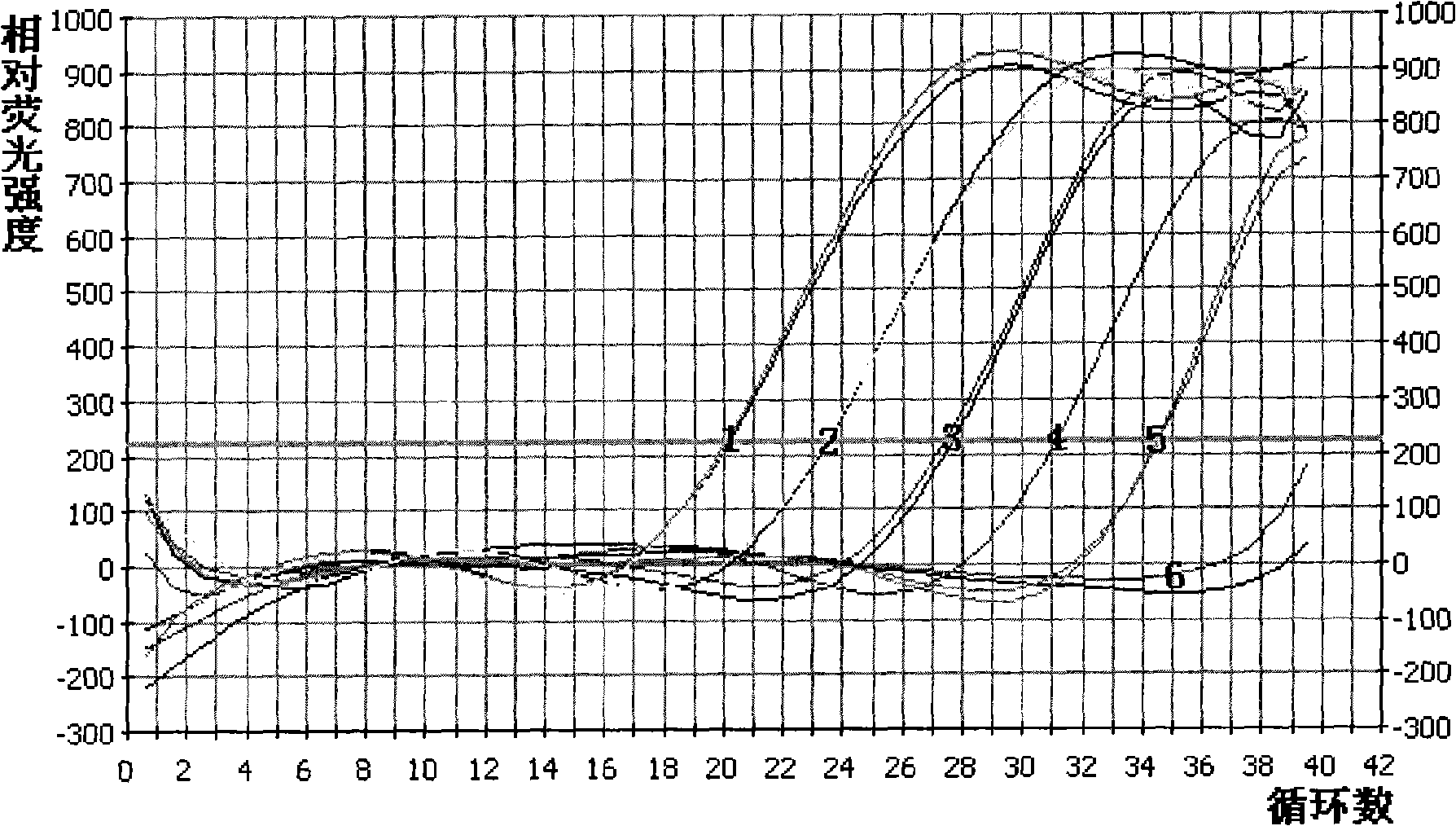
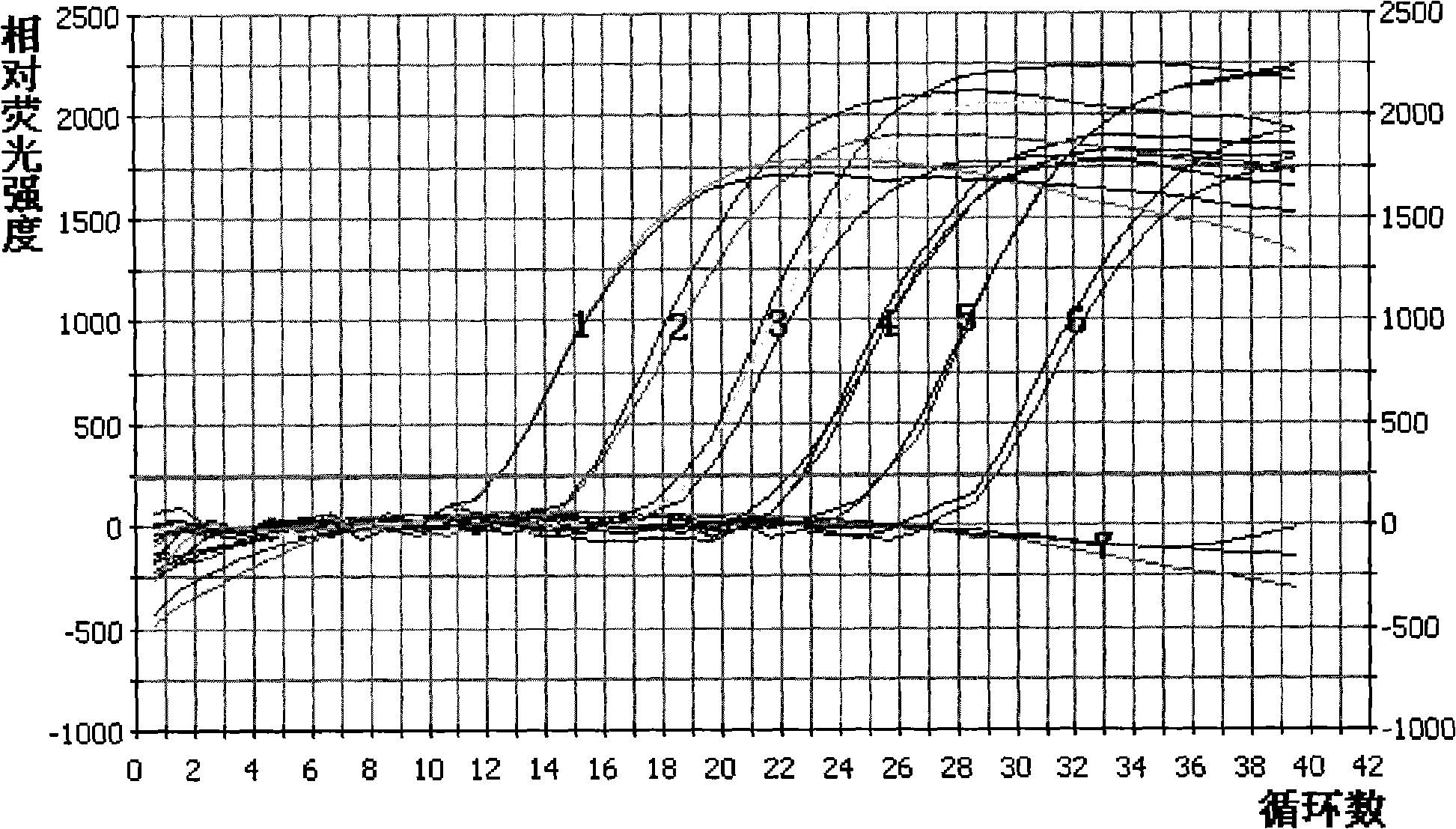
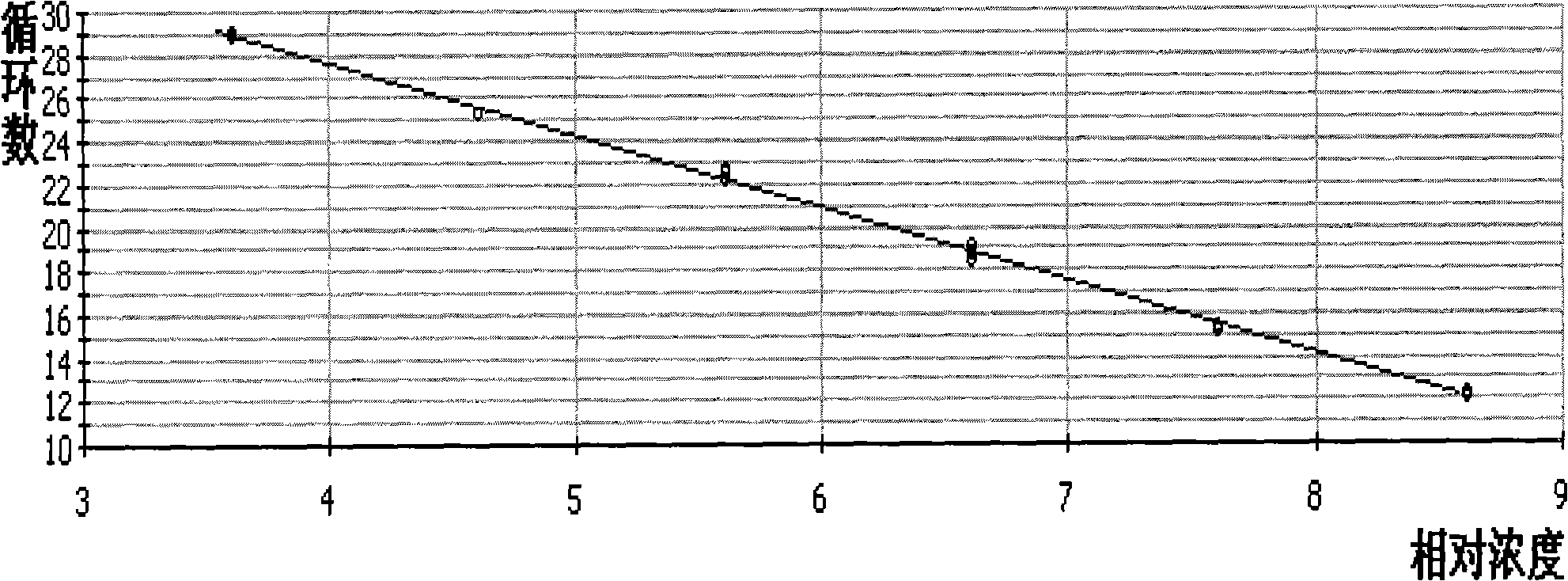
[0128] Make a 10-fold serial dilution of the standard plasmid containing the target gene whose copy number has been calculated to 10 copies / μl. Take the concentration as 10 6 、10 5 、10 4 、10 3 、10 2 、10 1 Copies / μl of the plasmid was used as a template for Real Time PCR. The lowest starting template concentration with product is the sensitivity of the Real Time PCR. According to the amplification curve analysis, the detection sensitivity of the Real Time PCR method is 10 copies / μl or 20 copies / 20 μl reaction system. See details figure 2 .
PUM
| Property | Measurement | Unit |
|---|---|---|
| Copy number | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com