Catalytically inactive proteins and method for recovery of enzymes from plant-derived materials
An inactive, xylanase-active technology, applied in the field of expression of mutant xylanase in microorganisms and yeast, can solve the problems of poor enzymatic activity, poor recoverability, sharp recovery, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0178] Example 1: Site-directed mutagenesis to change the catalytic nucleophile of xylanase from glutamic acid to alanine, producing a catalytically inactive protein
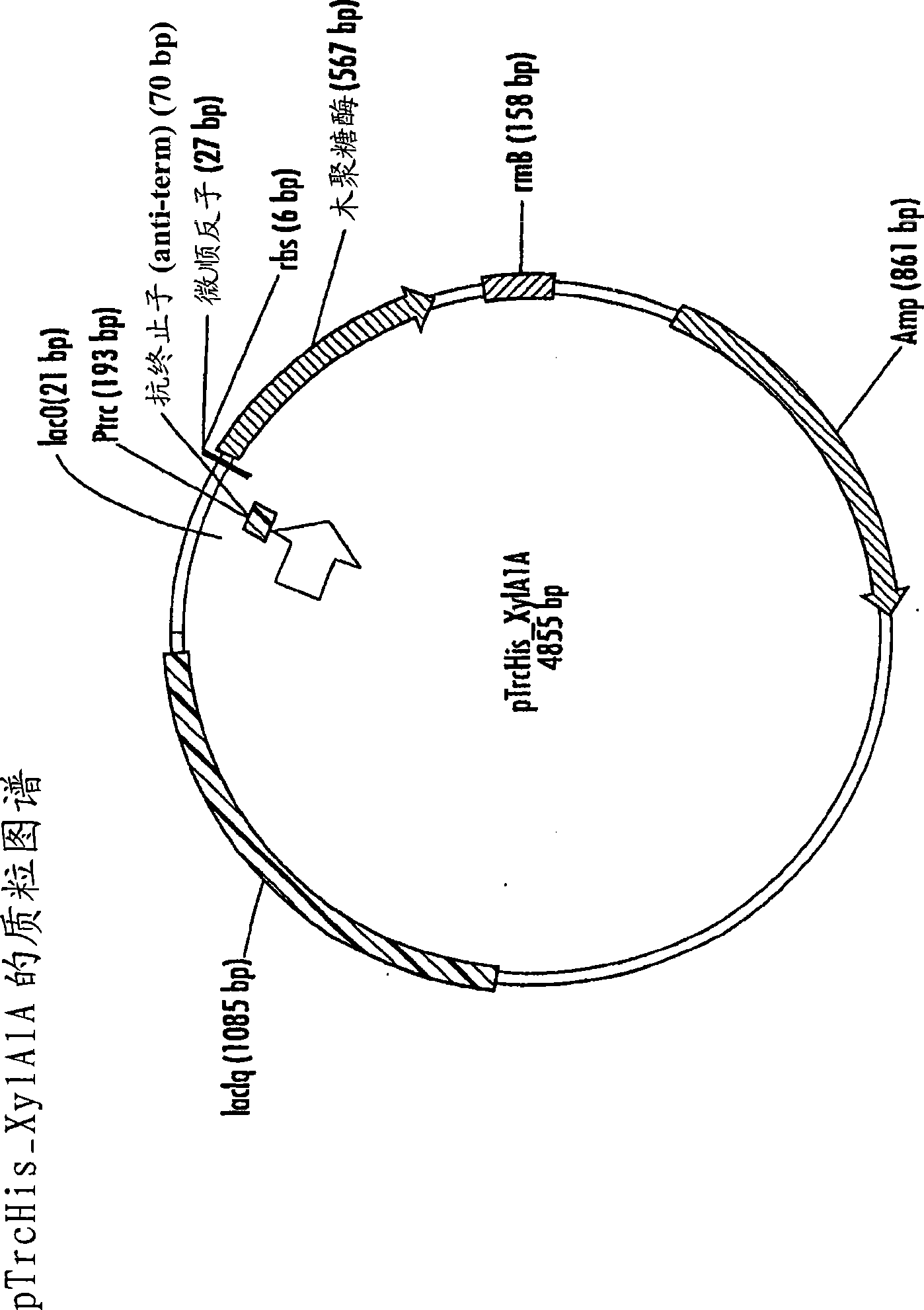
[0179] Xylanase Xy1A1A was identified by activity-based screening of libraries from environmental samples. The gene encoding wild-type Xy1A1A xylanase (SEQ ID NO. 3) was cloned into the bacterial expression vector pTrcHis. The vector is named pTrcHis_Xy1A1A, which is derived from figure 1 and represented by SEQ ID NO.114. To create this construct, the possible signal sequence of Xy1A1A was removed from the full-length gene sequence resulting in a truncated xylanase gene. Further, after inserting the truncated xylanase gene into pTrcHis, the open reading frame containing the xylanase gene contained five additional 5' terminal codon. Sequence alignment of the translated protein of the full-length Xy1A1A coding sequence and other glycosyl hydrolases family 11 xylanases in the literature shows that the 79th posi...
Embodiment 2
[0194] Example 2: Preparation of catalytically inactive xylanase proteins in bacterial expression hosts
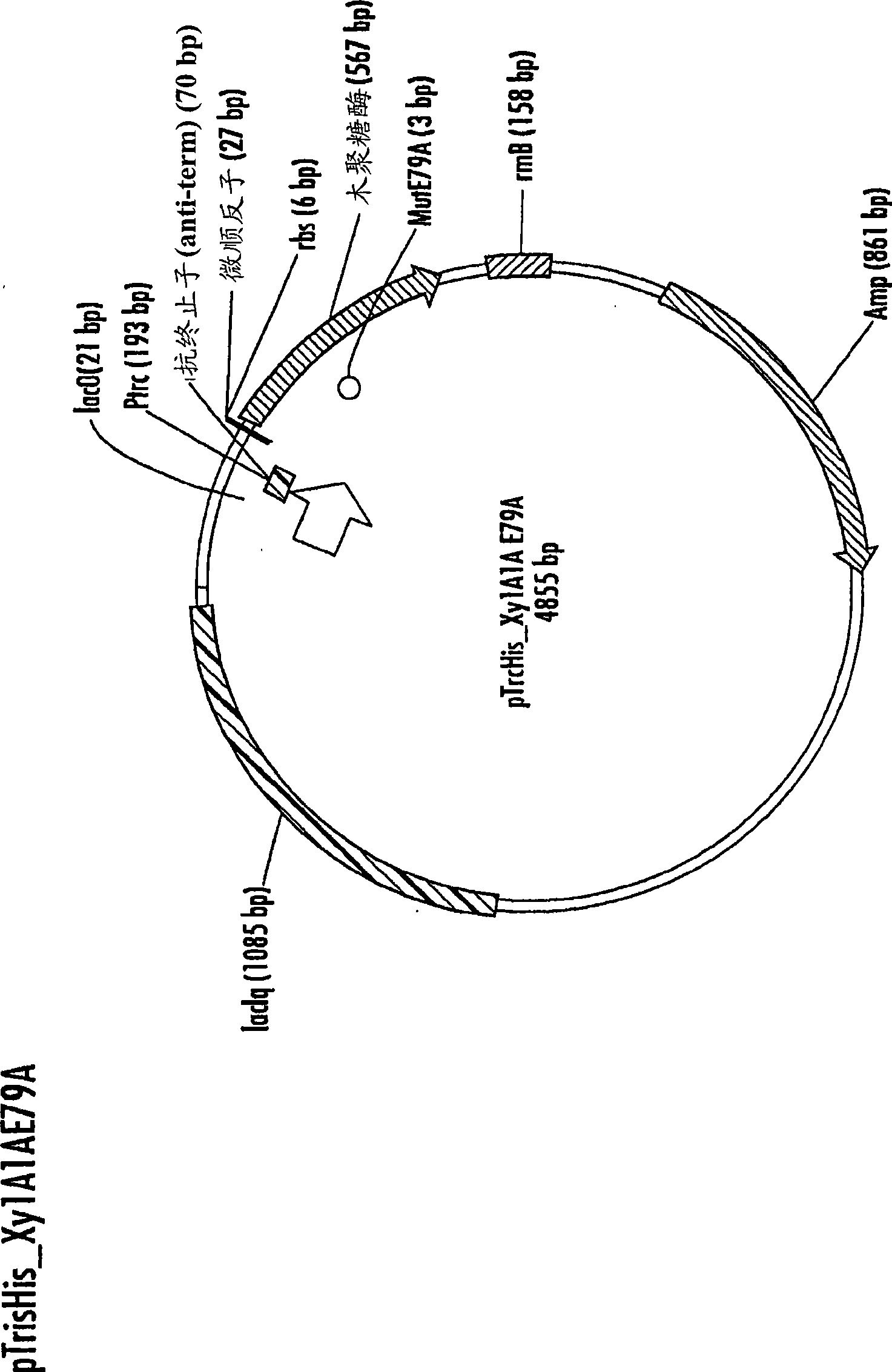
[0195] The pTrcHis_Xy1A1A_E79A vector was transformed into BL21 Star (pLysS) cells using standard techniques [Sambrook et al.] and spread to 100 μg / mL ampicillin (LB amp100 ) on Luria medium agar plates. Pick a single colony and inoculate into 3.5 mL containing 50 μg / mL ampicillin and 25 μg / mL chloramphenicol (TB amp50-chlor25 ) in Terrific medium, grown overnight at 37°C with continuous shaking. After culturing overnight, a portion of the culture was removed and stored as a frozen glycerol stock solution at -80°C.
[0196] From this glycerol stock, inoculate 20 mL of TB with a sterile loop amp50-chlor25 into a 250mL culture flask. Cultures were grown overnight at 37°C with shaking at 200-250 rpm. The next day, dilute 5 mL of the overnight culture to 1.5 L of TB amp50-chlor25 middle. The culture was grown at 37°C with shaking until the OD600 reached 0.6-1.0. Then, ...
Embodiment 3
[0197] Example 3: Preparation of expression constructs for the production of Xy1A1A_E79A in the yeast host Pichia pastoris
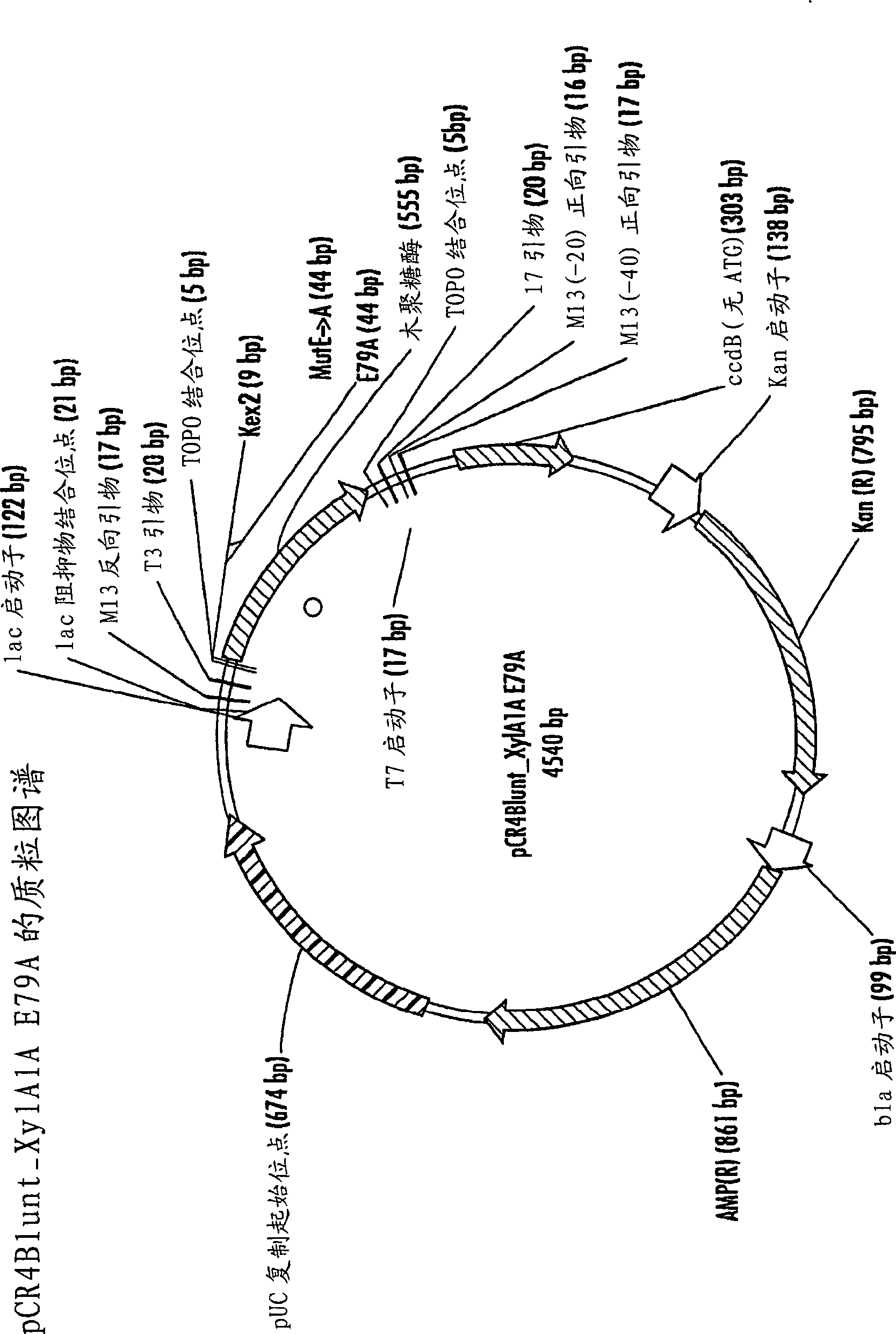
[0198] Construction of pCR4Blunt_Xy1A1A_E79A
[0199] Synthetic oligonucleotides (primers 3 and 4) and Pfu DNA polymerase (Stratagene, LaJol1a, CA) were used to PCR amplify the BD6002E79A gene from pTrcHis2-BD6002E79, and the parameters of the thermocycler used were set as follows:
[0200] Primer 3
[0201] 5'-TTTCCCTCTCGAGAAAAGAGCTTCGACAGACTACTGGCAAAATTGG (SEQ ID NO. 123)
[0202] Primer 4
[0203] 5'-TTTTCCTTTTGCGGCCGCCTATTACCAGACCGTTACGTTAGAGTAC (SEQ ID NO. 124)
[0204]
[0205] Primer 3 was designed to anneal at the codon corresponding to amino acid 5 on the xylanase-containing pTrcHis open reading frame. In addition, primer 3 added an XhoI restriction site and a Kex2 protease cleavage signal (Leu-Glu-Lys-Arg) in front of the mature xylanase coding sequence. Primer 4 contained a double stop codon after the xylanase gene. The BD6002E79A PCR...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com