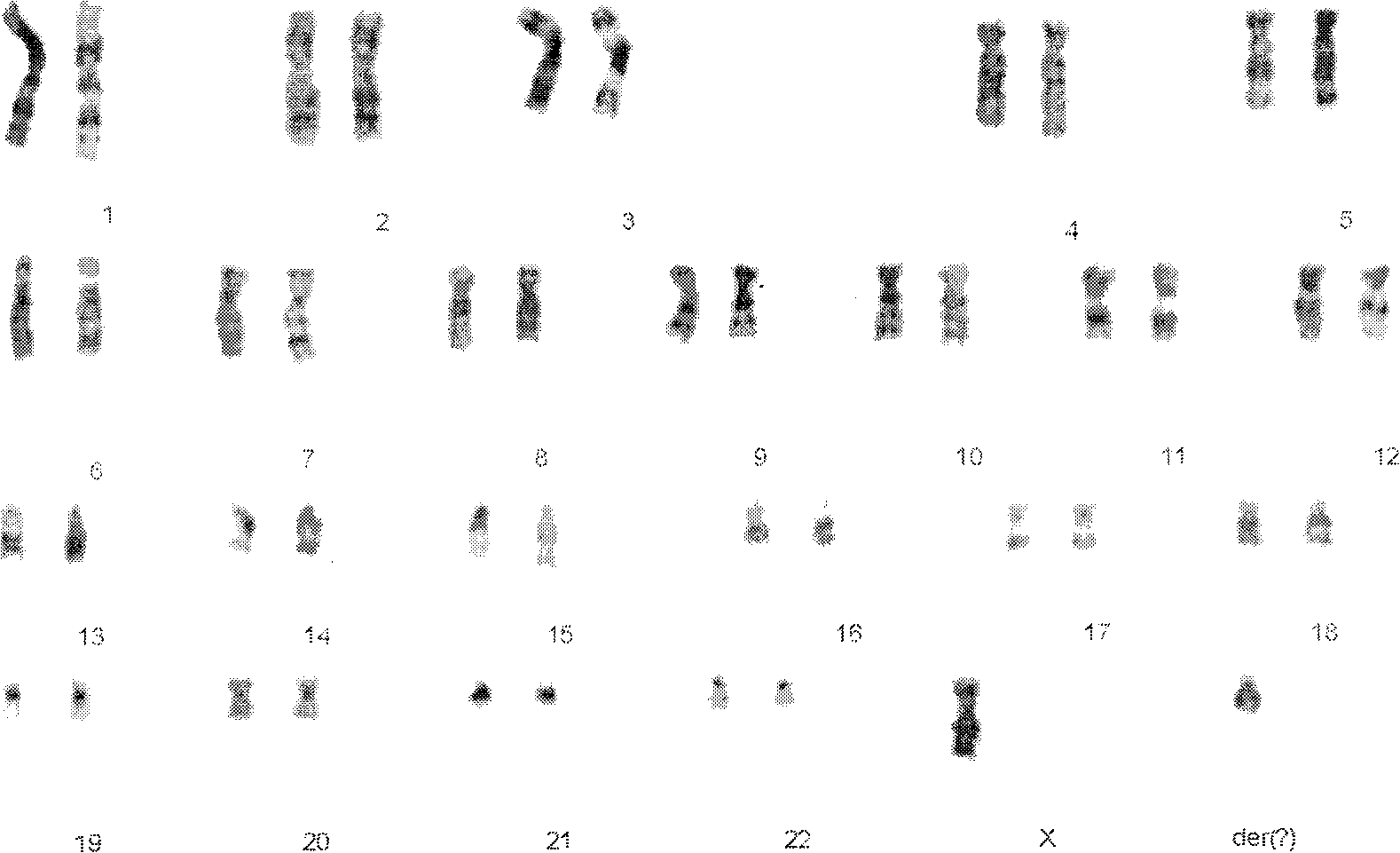Method for preparing birth defect target oligonucleotide microarray comparative genomic hybridization hybrid chip
An oligonucleotide and genome technology, which is applied in the field of preparation of birth defect-targeted oligonucleotide microarray comparative genomic hybridization chip, can solve the problems of limited targets, reduce the detection of CNVs of unknown clinical significance, Complete and reliable test results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Case information
[0027] A 37-year-old pregnant woman of Han nationality who had 3 deliveries and 0 births. At 26 weeks of pregnancy, the ultrasound examination revealed complete ECD in the fetus. The lover is 42 years old and denies consanguineous marriage, medical history of high blood pressure, diabetes, kidney disease, hepatitis, tuberculosis, etc., and family genetic medical history. G-banding chromosome analysis is difficult to determine the nature of the fetal Y chromosome, like the short arm of the X chromosome, so it is marked as a derived chromosome, and the fetal karyotype is: 46, X0, +der(?)( figure 1 ). The karyotypes of both parents were normal. The post-abortion examination revealed that the fetus was a male with six-digit deformity in front of the axis of the right hand, and no other abnormalities were found.
[0028] cell collection
[0029] Collect the remaining fetal cell suspension after the G-banded chromosome inspection and store at -20°C. Th...
Embodiment 2
[0047] Case information
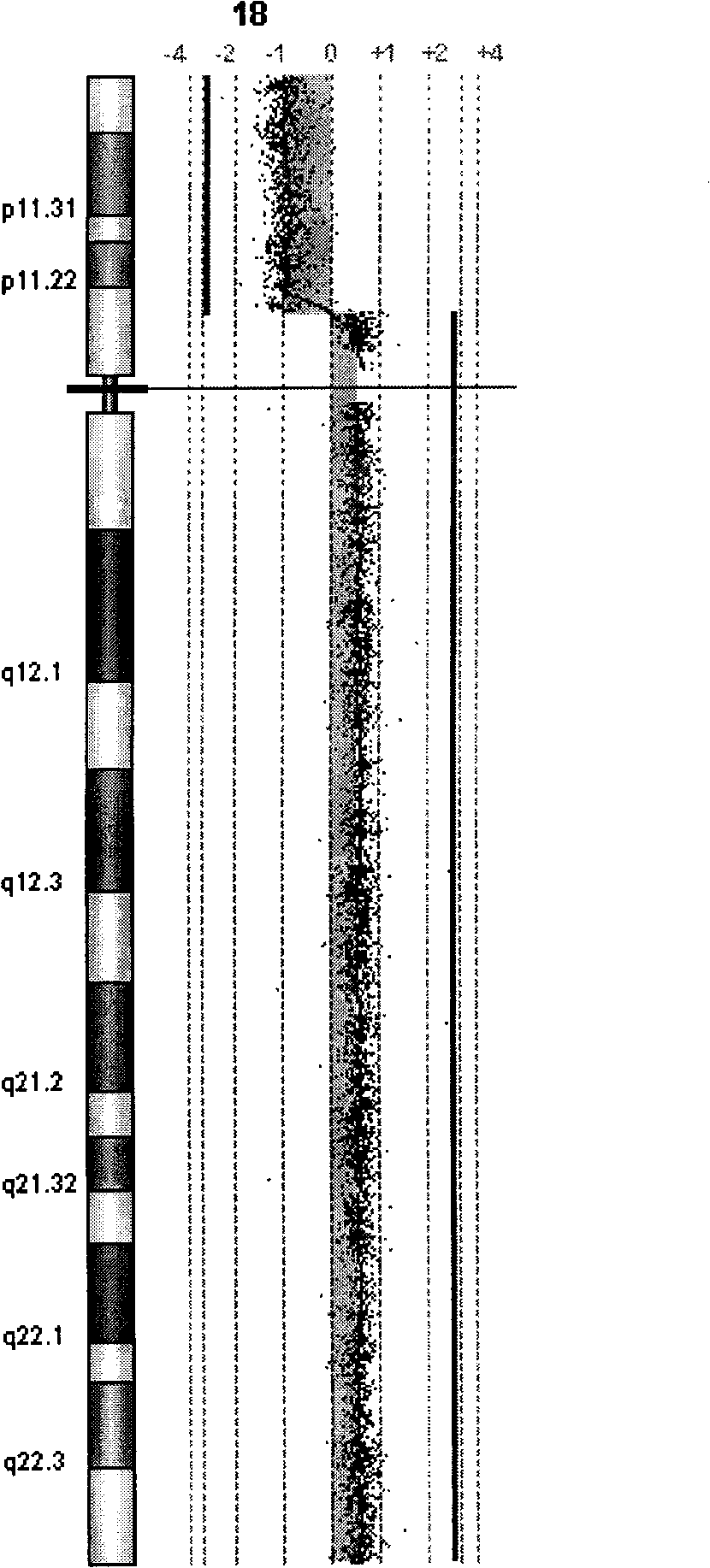
[0048] Han Huiling, 28 years old, Han nationality, 3rd pregnancy, 0 births, 26 weeks pregnant, was admitted to this hospital because of multiple fetal malformations found by ultrasound examination. Ultrasound found that the fetus had bilateral radial absence and deformed hands, short bilateral humerus, right foot varus, Taussing-Bing syndrome (ventricular septal defect, pulmonary artery overriding), bilateral choroidal cysts, and soft tissue thickening of the neck. He denied the marriage of consanguineous relatives, denied the history of hypertension, diabetes, kidney disease, hepatitis, tuberculosis, etc., and denied the history of family genetic diseases. The results of G-banded chromosome analysis are: 46, XY, -18, +der(?). FISH results indicated that the derived chromosome might be 18q isodicentric chromosome [idic(18)(p11→qter)], but the exact breaking point could not be determined yet. The karyotypes of both parents were normal.
[0049] cell...
Embodiment 3
[0070] Example 3 The surface of the slide is siliconized
[0071]1. Cleaning and silanization of glass slides. Soak the slides in 1mol / L NaOH for 2 hours. During the soaking process, put them on a shaker and vibrate them. After cleaning them with sterilized water, soak them in 1M HCL overnight. Then use Wash with sterile water and finally with methanol. Put the glass slide into 95% ethanol solution containing 1% glycidyl-propoxytrimethoxysilane (GOPS) for siliconization for 30 min, wash with ethanol solution, and dry.
[0072] 2. Coating with Poly-L-Lysine, immerse the silanized glass slide in 1:10 Poly-L-Lysine and PBS culture solution and shake at room temperature for 2 hours. After the reaction, wash the slide with double distilled water for 5 times, and then The slides were centrifuged at 500r / min for 5min in a centrifuge, and dried in a vacuum oven at 45°C.
[0073] 3. Surface activation, put the Poly-L-Lysine-coated glass slide in the previous step into the activation ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com