Fusion protein containing single-stranded DNA binding protein, expression and purification methods thereof
A technology of fusion protein and binding protein, which is applied in the field of expression and purification of fusion protein, which can solve the problems of high cost, low purification efficiency, and long time required
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Example 1: Construction and expression of expression plasmid vectors pSSB-B1, B2, B3 and B4
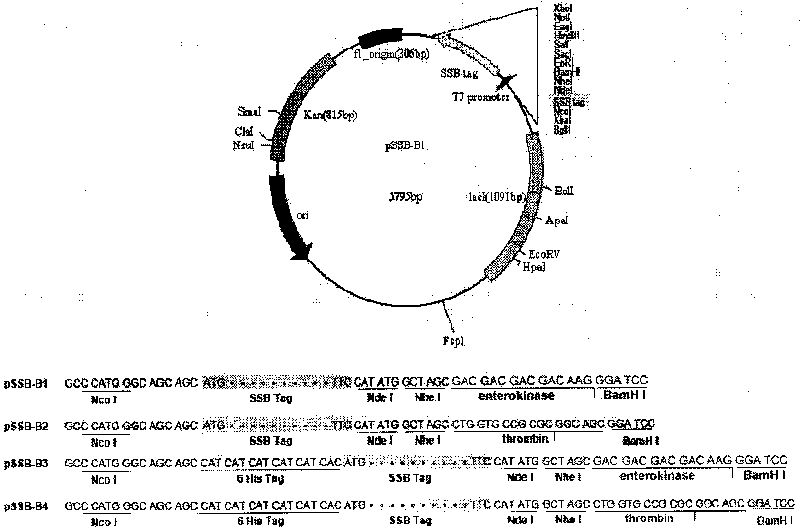
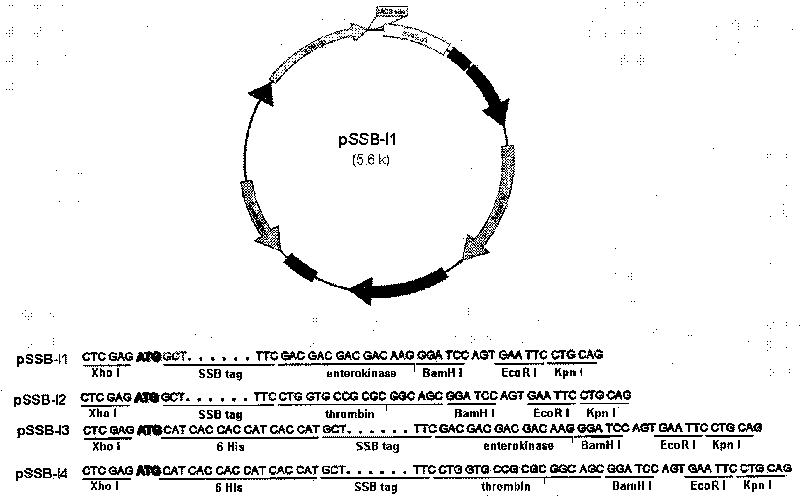
[0051] Primers were designed for polymerase chain reaction (PCR), and the DNA fragment encoding T7SSB was obtained from the phage T7 genomic DNA, and inserted into the Nco1 and BamH1 sites of the expression vector pET28a to obtain the expression vector pSSB-B1. In the pSSB-B1 vector, an enterokinase-recognized junction fragment is located between the SSB tag and the BamH1 site. Expression plasmid vectors pSSB-B2, B3 and B4 were similarly constructed. In these four plasmids, the cleavage site for SSB and enterokinase or thrombin is located between the NcoI and BamHI sites of the pET28a vector. Additionally, in the vectors pSSB-B3 and pSSB-B4, a hexa-histidine tag was placed just at the N-terminus of SSB. For physical maps and multiple cloning sites see Figure 1A . This series of plasmid vectors is used to express fusion proteins with SSB or 6His-SSB tag in E.coli.
Embodiment 2
[0052] Embodiment 2: Construction of expression plasmid vector pSSB-Y1 and Y2
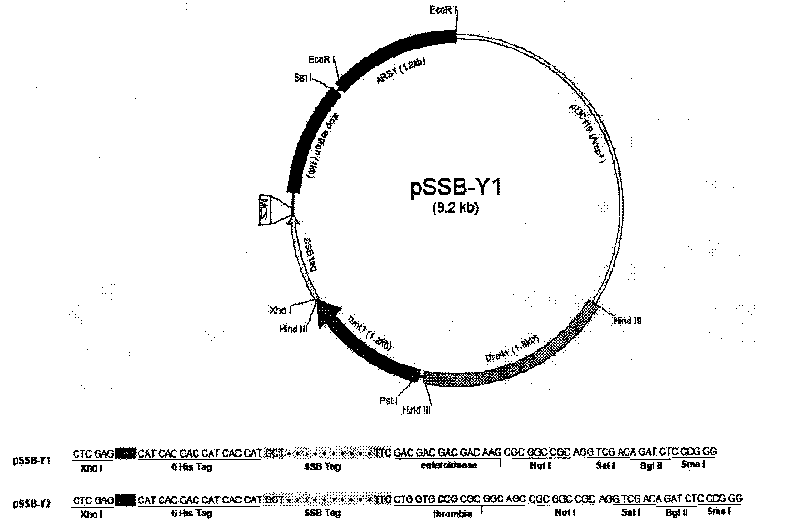
[0053] Primers were designed for polymerase chain reaction (PCR), and the DNA fragment encoding T7SSB was obtained from the phage T7 genomic DNA, and inserted into the XhoI and NotI sites of the expression vector pSLF1072 to obtain the expression vectors pSSB-Y1 and Y2. In pSSB-Y1, the cleavage site of enterokinase is located between SSB tag and NotI site; in pSSB-Y2, the cleavage site of thrombin is located between SSB and NotI site. Additionally, in the vectors pSSB-Y1 and pSSB-Y2, a hexa-histidine tag was placed just at the N-terminus of SSB. For physical maps and multiple cloning sites see Figure 1B . This series of plasmid vectors is used to express fusion proteins with SSB or 6His-SSB tag in yeast.
Embodiment 3
[0054] Example 3: Overexpression of fusion proteins 6His-SSB-Sap1 and SSB-Fen1 in E.coli and purification by single-stranded DNA cellulose chromatography
[0055] Sap1 is an essential protein related to the initiation of DNA replication, which exists in large quantities in fission yeast, with a molecular weight of about 30kDa. The DNA fragment of the sap1 gene was amplified from the fission yeast genomic DNA by PCR reaction, and inserted into the BamHI and HindIII sites of the pSSB-B4 vector. The recombinant plasmid encoding the fusion protein 6His-SSB-Sap1 was transformed into E.coli BL21(DE3)pLysS cells to obtain a single clone. Single clones were cultured overnight in 50 mL LB medium containing 20 μg / ml kanamycin. The overnight bacterial solution was transferred to fresh medium at a ratio of 1:50 to 1:100, and cultivated to OD at 37°C 590 =~0.3. Then add IPTG to 0.1 mM and induce for 3.5 hours. The cells were then centrifuged and suspended in buffer A (50 mM Tris-HCl, p...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com