Small RNA (Ribonucleic Acid) quantitative detecting method and reagent kit
A quantitative detection method and quantitative detection technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems of low detection sensitivity, high false positive rate, and the inability to detect short fragments of small RNA, etc. Achieve the effect of high detection sensitivity, good specificity and cheap kit
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0046] Example 1: Detection of miRNA expression in cells
[0047] Take the detection of hsa-let-7a, hsa-miR-10b expression in A549, HeLa, HEK293 cells as an example.
[0048] 1. Related primer design
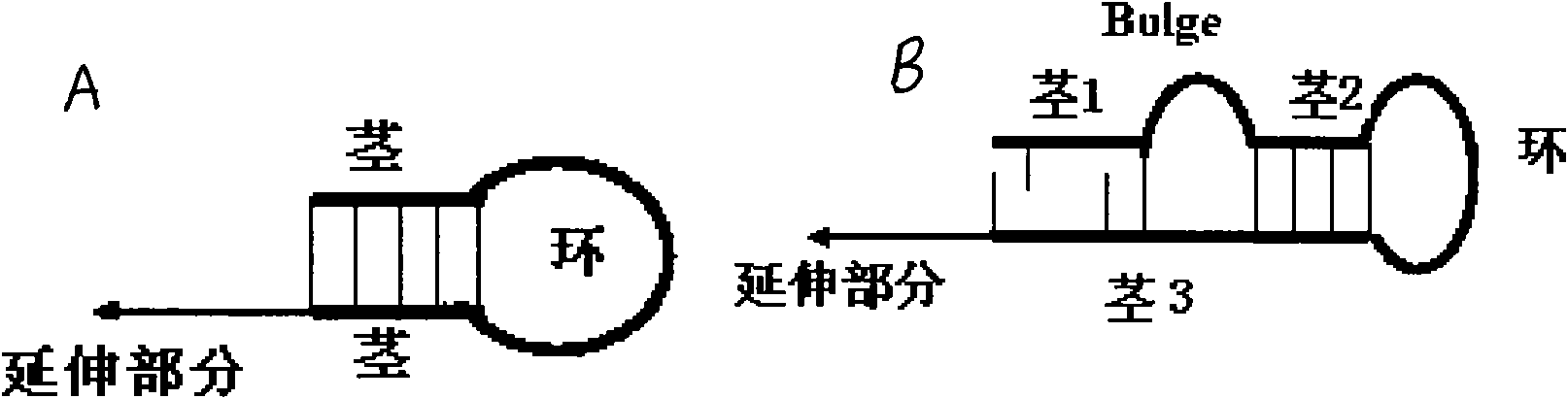
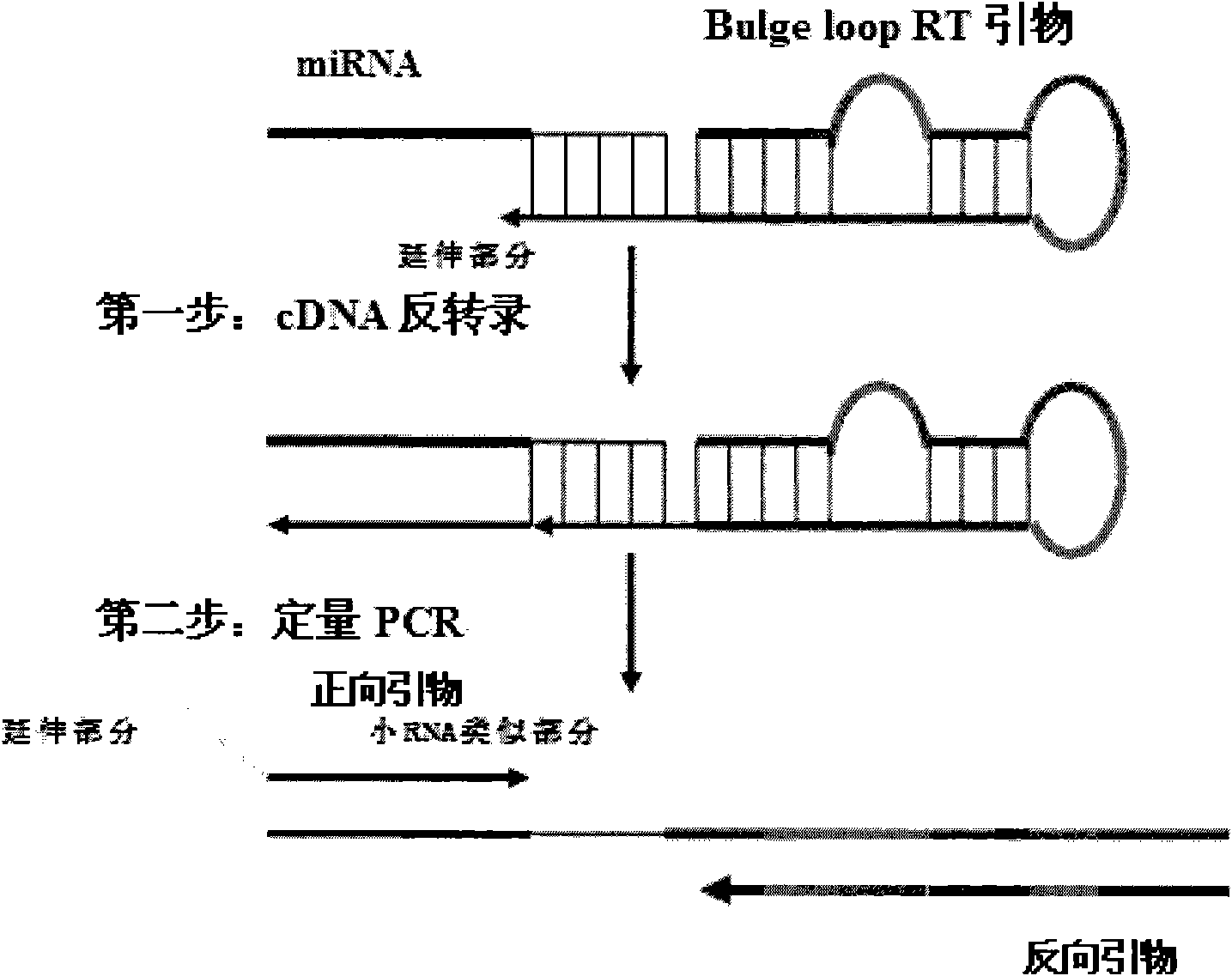
[0049] 1), RT primers with Bulge loop structure:
[0050] hsa-let7a-RT1: (SEQ. NO: 1) GCACTTCAGTGTCGTGGTCAGTGACGGCAATTTGAAGTGCAACTATAC
[0051] hsa-miR10b-RT1: (SEQ. NO: 2) GCACTTCAGTGTCGTGGTCAGTGACGGCAATTTGAAGTGCCACAAATT
[0052] 2), PCR forward primer
[0053] hsa-let7a-F1: CAGACGACCATCAGTGAGGTAGTAGGTTGA (SEQ. NO: 3)
[0054] hsa-miR10b-F1: CAGACGACCATCAGTACCCTGTAGAACCGAA (SEQ.NO: 4)
[0055] 3), universal reverse primer
[0056] G-R1: GCACTTCAGTGTCGTGGTCAGTGACGGCAATT (SEQ. NO: 5)
[0057] 2. cDNA preparation
[0058] A549, Hela, and HEK293 cells were used as materials respectively, and the cells were lysed with Trizol from Invitrogen Company, and the total RNA of the cells was extracted by adding chloroform and isopropanol according to routine operations, and hsa-let7...
Embodiment 2
[0062] Detection of miRNA expressed in human secretions, blood, and semen
[0063] Human secretions include saliva, vaginal secretions, sweat, etc. Saliva, semen and other liquid samples were added to Trizol at a ratio of 1:3 to extract RNA, and vaginal secretions were transferred to EP tubes with cotton swabs and added to Trizol for RNA extraction. Blood samples were collected in EDTA anticoagulant tubes, and a part was taken as a whole blood sample. The anticoagulated blood was centrifuged at 4000 rpm for 10 minutes at 4 degrees, and the upper layer was plasma. Plasma samples were added to Trizol at a ratio of 1:3 to extract RNA. The primer sequence is shown in Example 1, and the miRNA detection result is as follows Figure 5 , the test results show that this method can be used for the detection of small RNA in human secretions.
Embodiment 3
[0065] Detection of miRNA expressed in blood
[0066] Taking the detection of miR-150 in human blood samples as an example, U6 was used as an internal reference, and leukocytes were collected by centrifugation after blood collection. The RNA extraction of leukocytes and the detection process of miRNA were the same as in Example 1. The sequences of related primers for miR-150 and U6 were as follows:
[0067] name
[0068] The test results show that this method can detect the expression of miRNA from blood cells ( Figure 6 ).
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com