Nucleotide sequence fragments that promote high-efficiency expression of foreign proteins in cho cells
A nucleotide sequence and high-efficiency expression technology, applied in the field of nucleotide sequences derived from CHO cells, can solve problems such as insufficient expression, poor predictability of protein production, and unstable long-term expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1: Obtaining and Sequence Identification of KH Sequence
[0033] Take about 1×10 cultured CHO cells 7 One, using the genome extraction kit to obtain the CHO genome, and then design the specific upstream primer KHF: 5'>TGCTTTGAATGTTCAAGTAGGATCTAATGTACATCATGAG<3';
[0034] The PCR reaction system is: CHO genomic DNA 2ul, dNTP2ul, KHF2ul, KHR2ul, LA-taq 0.5ul, 10×LA-taq Buffer 2ul, H 2 O 9.5ul, total volume 20ul;
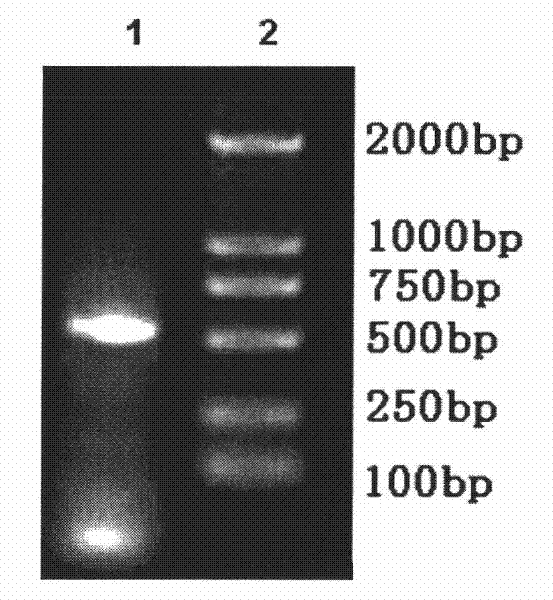
[0035] The PCR reaction program was as follows: 10 min at 94°C, 30 sec at 94°C, 30 sec at 60°C, 45 sec at 72°C, a total of 32 cycles, and finally 10 min at 72°C. For the results of the PCR reaction, see figure 1 .
[0036] The obtained PCR product was directly loaded into the pMD 19-T vector for sequencing identification. The sequencing result is shown in SEQ ID NO.1.
Embodiment 2
[0037] Embodiment 2: Apply KH sequence to make luciferase in CHO / dhfr - highly expressed in cells
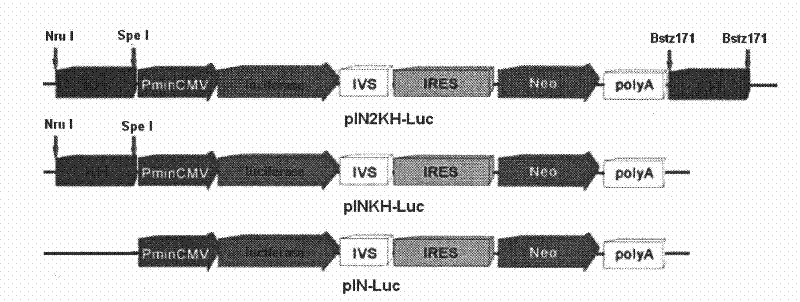
[0038] Take the pIRESneo3 plasmid, insert the nucleotide sequence into the upstream, downstream and upstream of the exogenous gene transcript, and construct two different vectors, named pIN-KH and pIN-2KH respectively; then insert luciferase into The multiple cloning sites were constructed as recombinant expression vectors named pIN-KH-Luc and pIN-2KH-Luc respectively, such as figure 2 . In addition, the control plasmid pIN-Luc was constructed again.
[0039] Using liposome transfection method, pIRESneo3, pIN-Luc, pIN-KH-Luc and pIN-2KH-Luc were transfected into CHO / dhfr-cells respectively, and an internal reference plasmid pRL-TK was transfected at a ratio of 1 / 10 The transfection was carried out on a 24-well cell culture plate, using different amounts of liposomes and DNA. For the operation method, refer to the instruction manual of Lipofectamine2000 liposome reagent provi...
Embodiment 3
[0048] Example 3: Using KH sequence to make FGF2 highly expressed in CHO / DG44
[0049] 2.1 Construction, identification and large-scale preparation of recombinant expression vectors
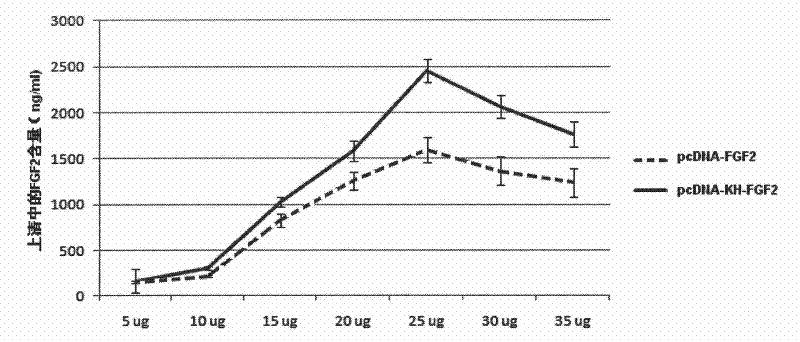
[0050] Using the nucleotide sequence of Fibroblast Growth Factor 2 (FGF2) as a template, FGF2 was amplified by PCR, double digested with Nhe I and HindIII, and the eukaryotic expression vectors pcDNA3.1 and pcDNA3.1-KH were taken, Digested with Nhe I and HindIII, connected with T4 ligase and transformed into competent Escherichia coli DH5α, spread on LB-AMP+ plate, 16-18 hours at 37°C, screened and picked 3-5 single colonies, in 5ml The LB-AMP+ medium was expanded, the plasmid was extracted, the recombinant was identified by two methods of enzyme digestion and PCR amplification of the target sequence, and the correct recombinant expression vector was obtained by sequencing and identification, which were named pcDNA3.1-FGF2 and pcDNA3 respectively. 1-KH-FGF2. The recombinant strain was amplified...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com