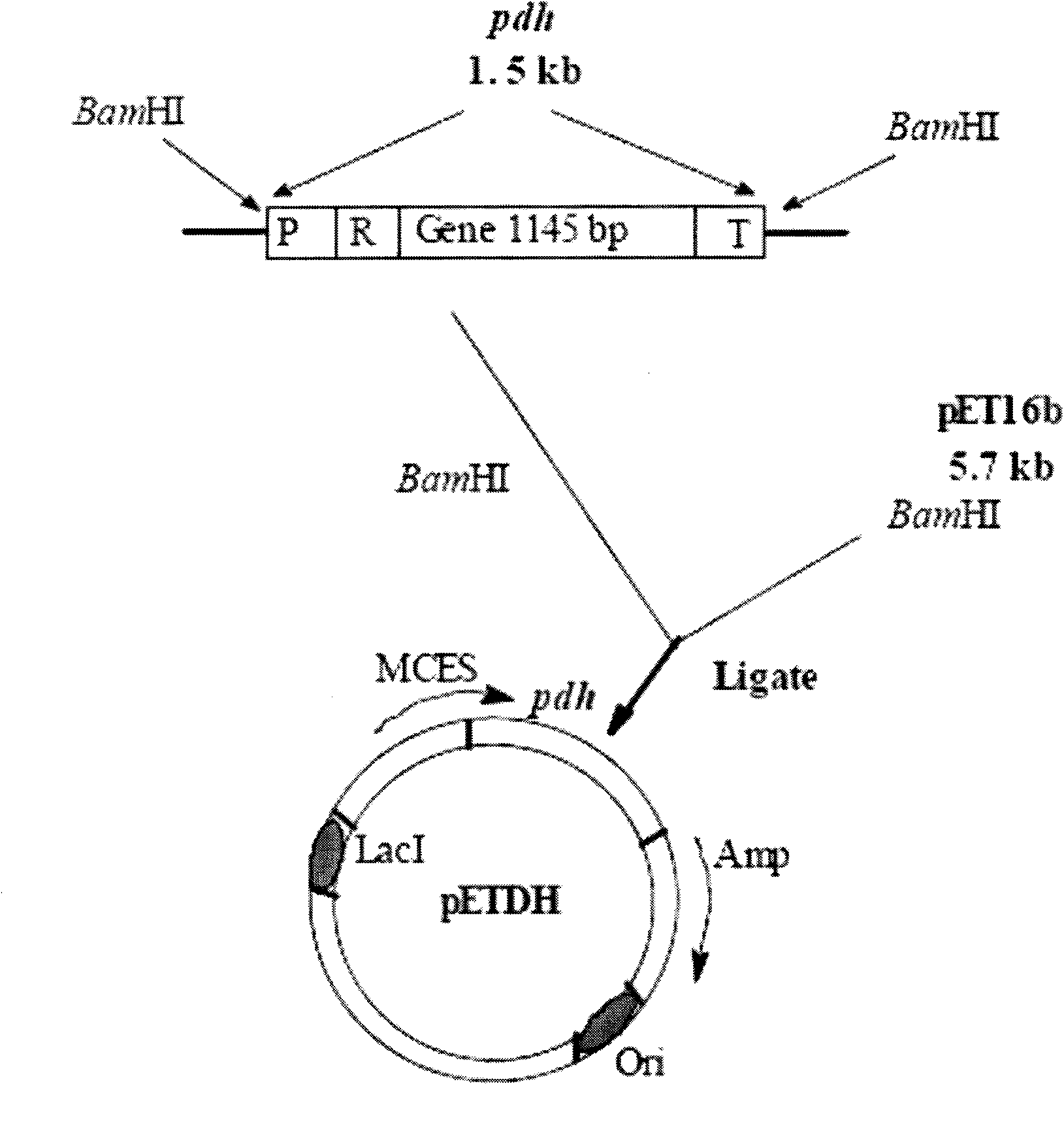
Technical method for preparing phenylalanine hydroxylase
A technology of phenylalanine carboxylase and phenylalanine dehydrogenase, which is applied in the field of high-yield L-phenylalanine dehydrogenase, can solve the problems such as tedious and time-consuming experimental purposes of the purification method, and achieves high yield, The effect of simple production method
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1 Preparation of chromosomal DNA containing L-phenylalanine dehydrogenase gene
[0028] Inoculate the following media with Bacillus sphaericus: 2g / l L-phenylalanine, 5g / l K 2 HPO4, 10g / l digested protein, 1g / l NaCl and 0.5g / l MgSO 4 ·7H 2 O, pH7.0. Shake culture at 30°C for about 10 hours until OD616=1.1.
[0029] Bacteria solution was centrifuged to obtain 10g wet bacteria, and chromosomal DNA was extracted by known methods, that is, bacteria were suspended in 40ml buffer solution TEN (10mM Tris-HCl, pH 7.6, 1mM EDTA and 10mM NaCl), centrifuged, collected cell. Then the collected cells were suspended in 20ml buffer SET (20% sucrose, 50mM Tris-HCl, pH 7.6 and 50mM EDTA), 10mg lysozyme was added, and incubated at 37°C for 30min. After confirming the formation of bacterial cells under a microscope, 10 ml of buffer solution SET and 0.25 g of SDS were added to the suspension to lyse the bacterial cells. Lysates were extracted with phenol / chloroform, and DNA wa...
Embodiment 2
[0031] Embodiment 2 Chromosomal DNA is inserted in the vector
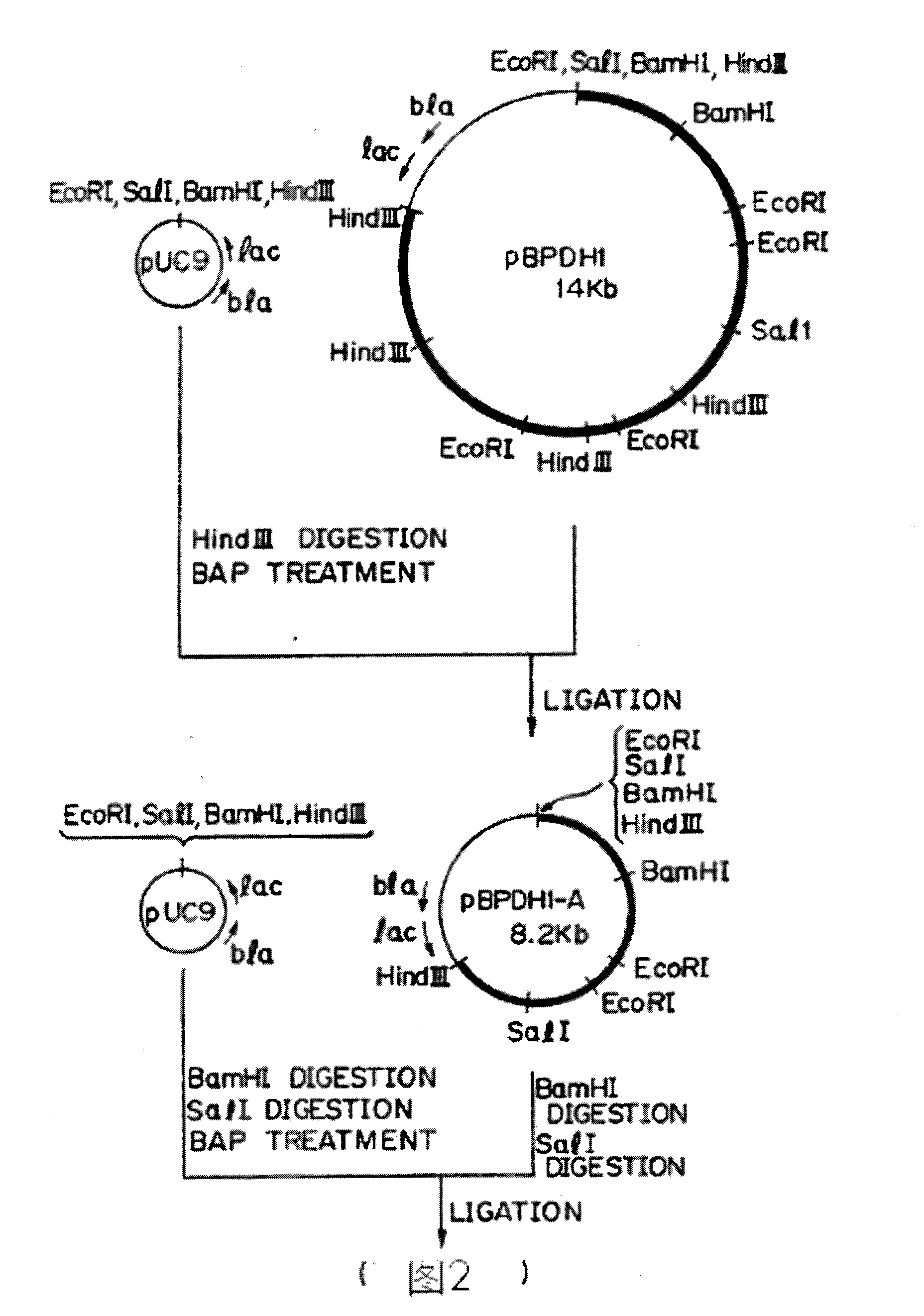
[0032] Plasmid pUC9 was used as a vector, and 10 μg of pUC9 was digested with Hind III at 37°C. Then, it was treated with calf thymophosphatase at 37°C for 2 hours, and then with ethanol for 1 hour.
[0033] The treated solution was subjected to agarose electrophoresis (1%), and the 2.7kb DNA fragment was isolated from pUC9 by electroelution method, extracted with phenol / chloroform, and then the carrier DNA was precipitated with ethanol, and dissolved in 20 μl sterile in the water.
[0034] On the other hand, as described in Example 1, 50 μg of chromosomal DNA was digested with 50u Hind III at 37°C, extracted with phenol / chloroform, and then precipitated with ethanol for 16 hours, and the obtained DNA was dissolved in 50 μl sterile water . The 0.5 μg vector DNA and 2 μg chromosomal DNA were mixed, and treated with T4 DNA ligase at 12.5° C. for 16 hours in the presence of ATP and dithiothreitol to form a recombi...
Embodiment 3
[0036] The screening of embodiment 3L-phenylalanine dehydrogenase positive bacteria
[0037] Transfect the above transformation mixture onto a nitrocellulose membrane. When each piece of nitrocellulose membrane reaches 500-1000 cloned recombinants, stick the nitrocellulose membrane on an LB plate containing 50 μg / ml ampicillin and incubate at 37°C 16 hr, followed by counterstaining with fresh nitrocellulose membranes incubated at 37°C for 3 hr.
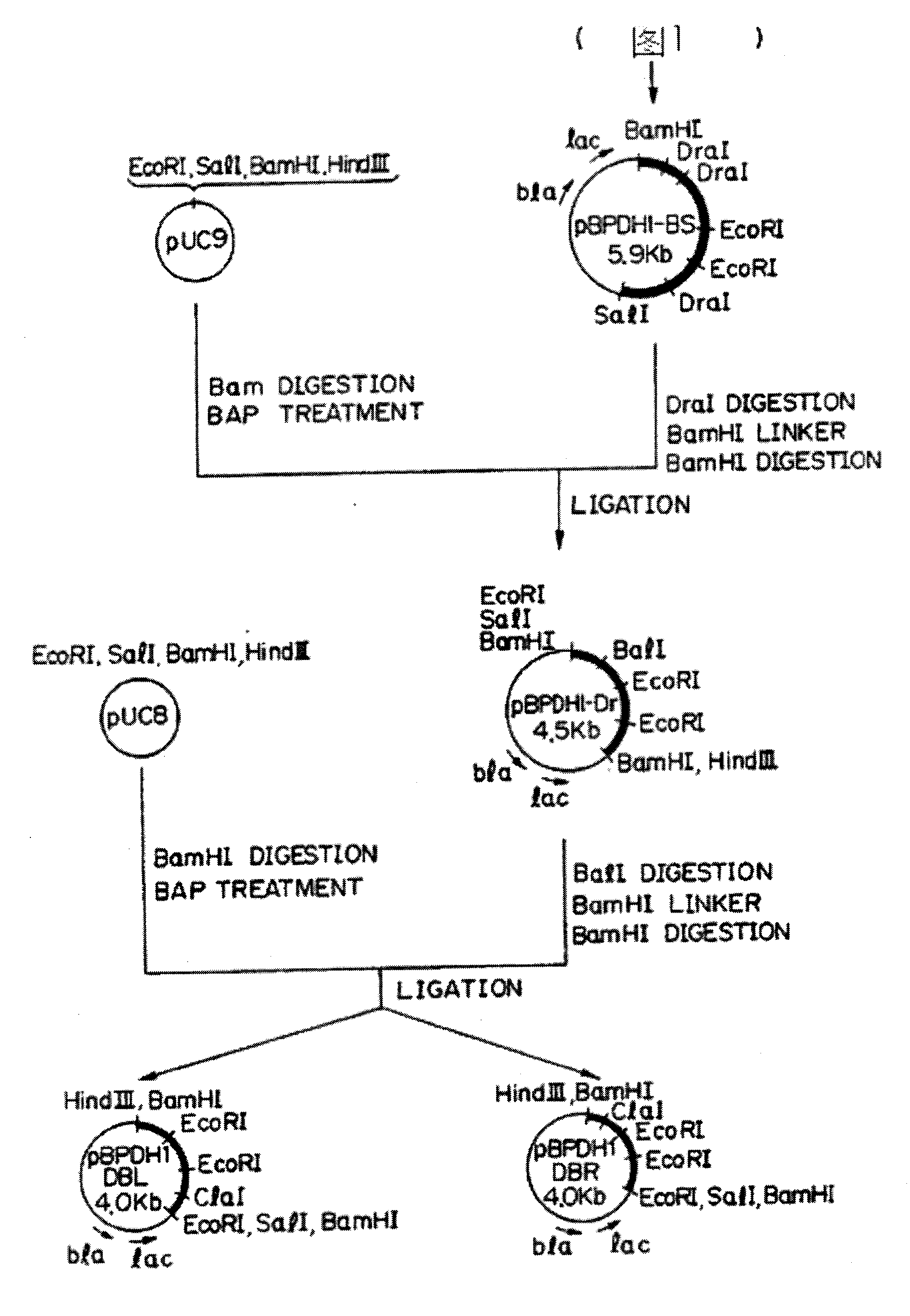
[0038] The nitrocellulose membrane was immersed in lysate (5 mg / ml lyase, 10 mM Tris-HCl, pH 8.5, 1.5 mM NAD+, 0.8 mMINT, and 0.32 mM phenazine dimethyl sulfate), and dark red transformants were selected. One L-phenylalanine dehydrogenase-positive colony provides about 2000 ampicillin-resistant transformants. Plasmids were extracted from L-phenylalanine dehydrogenase-positive colonies, and the recombinant plasmid from pUC9 was used as a vector, which was called pBPDH1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com