Rice rhizosphere burkholderia and application thereof in prevention and control of rice sheath blight disease
A technology of rice sheath blight and Burkholderia, applied in the direction of application, bacteria, fungicides, etc., to achieve low protease activity, high safety in use, and good inhibitory effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] Example 1 Isolation and Purification of Bacterial Strains
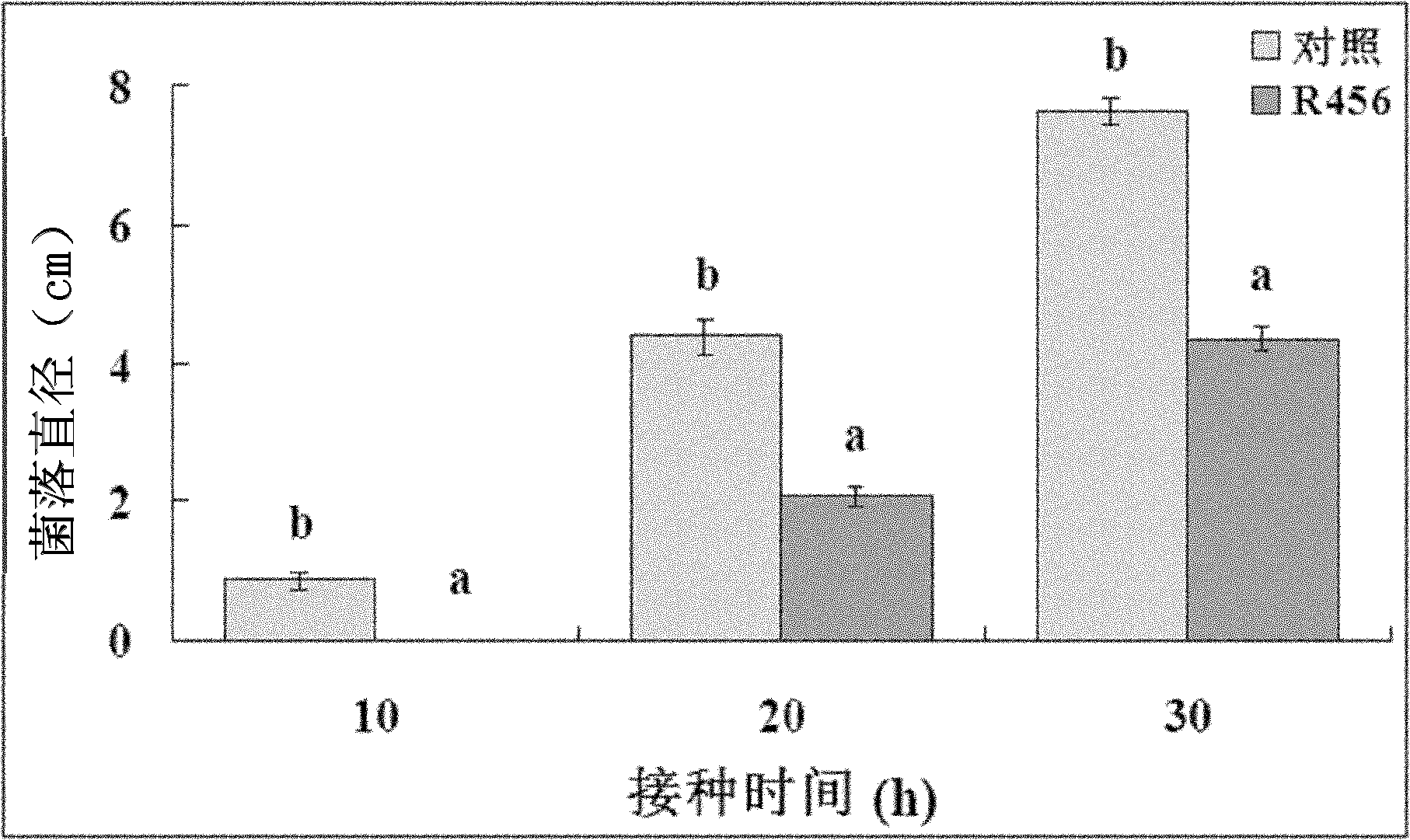
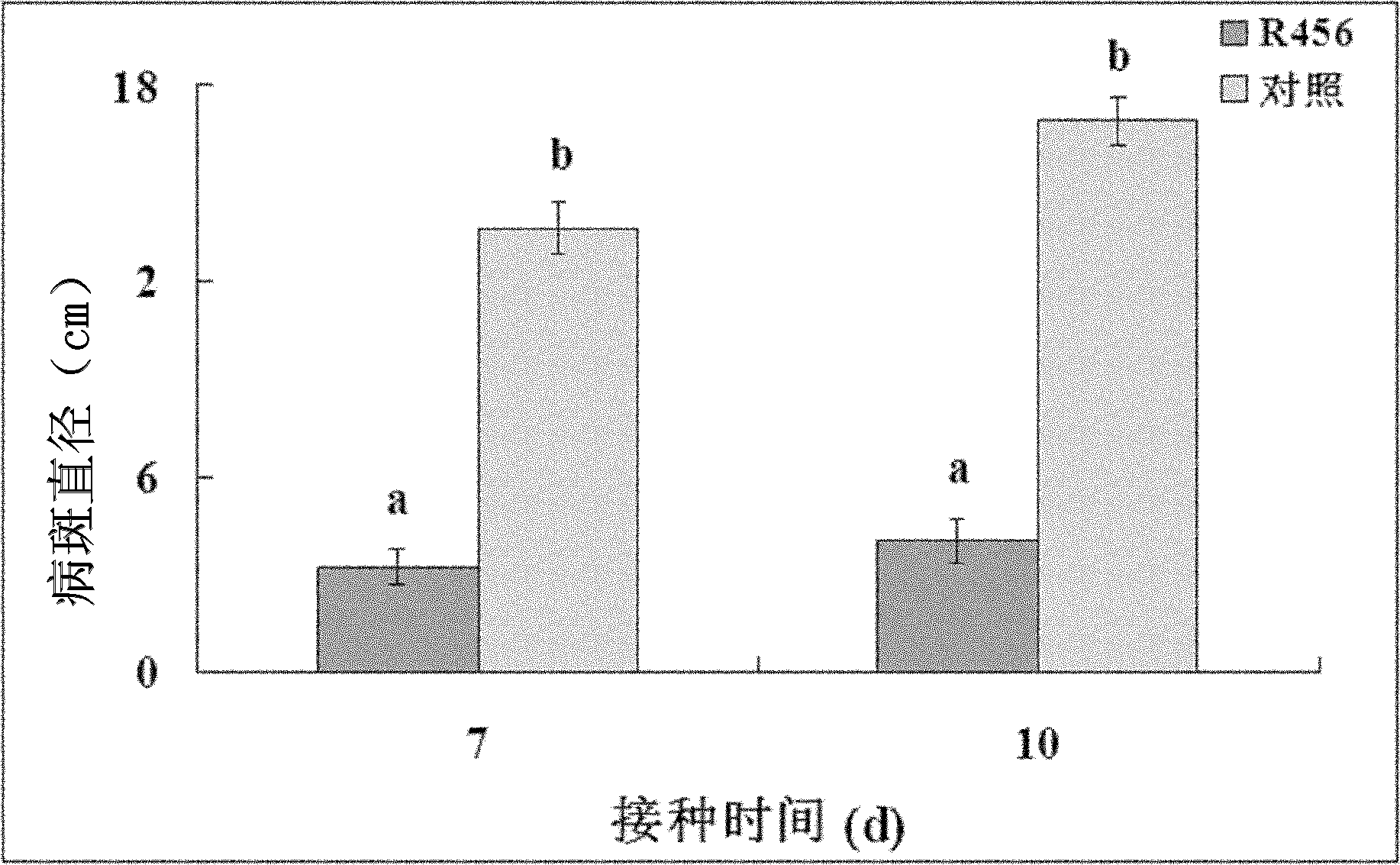
[0018] The 10-20cm surface soil around the rice rhizosphere was collected from a paddy field in Hangzhou City, Zhejiang Province, put into a sterile polyethylene plastic bag, and sealed. Weigh 10g of soil, add it into a triangular flask with 90ml sterile water to prepare a suspension, shake it at 180r / min for 30min, then draw 5ml from the resulting suspension and add it to a triangular flask with 45ml sterile water, and gradually dilute to Until the 5th triangular flask, get 10 -1 、10 -2 、10 -3 、10 -4 and 10 -5 diluent. Take 100μl 10 -4 and 10 -5 The dilutions of the above-mentioned nutrient agar medium were spread on the nutrient agar medium, and each dilution was repeated 3 times. After being cultured in a 30°C constant temperature incubator for 48 hours, a single colony with different cultural characteristics was picked and purified as the test strain. . Through the bacteriostatic diagram experiment...
Embodiment 2
[0020] Example 2 Identification of fatty acids
[0021] The specific test method is as follows:
[0022] 1. Place the strain at 28°C and culture it on trypticase soybean agar medium (containing: tryptone 15g, soybean peptone 5g, sodium chloride 5g, agar 15g, water 1000ml) for 24h;
[0023] 2. Use the MIDI fatty acid identification system to determine the fatty acid composition of the above strains;
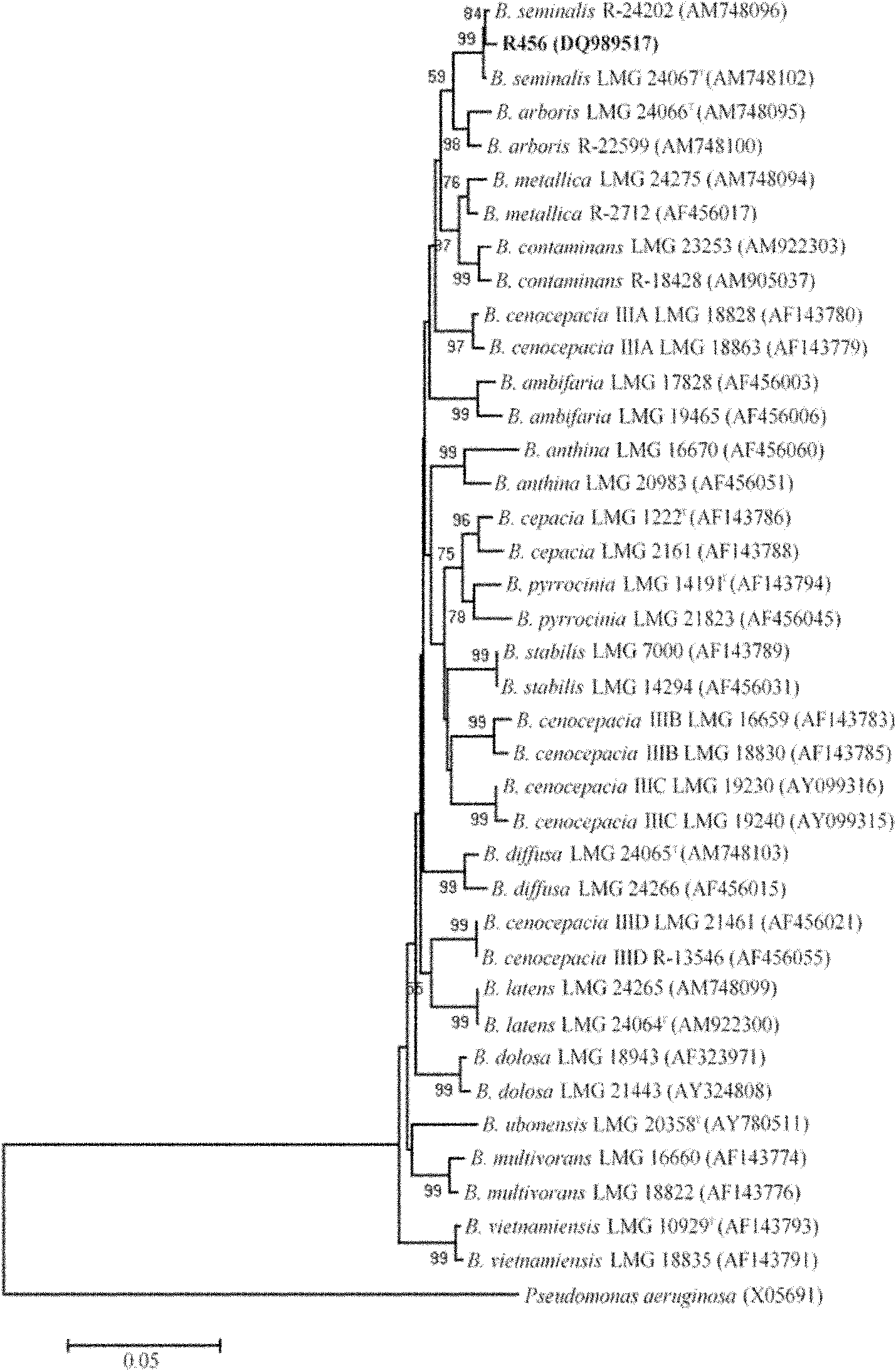
[0024] 3. According to the comparison of the determined fatty acid composition of the strain and the MIDI identification database TSBA50 version 5.00, the strain was identified.
[0025] According to the instruction manual of the microbial fatty acid identification system, a similarity value of 0.6 or higher indicates a good match, while a similarity value of 0.4-0.6 indicates that the two are not the same species. Compared with the species in the aerobic bacteria database in the microbial fatty acid identification system, it was found that the similarity value of this strain to...
Embodiment 3
[0026] Example 3 Biolog Carbon Source Utilization Identification
[0027] The biolog method was used to detect the utilization rate of the carbon source by the strain, and the specific method was as follows: the strain was placed in the tryptic casein soybean agar culture solution, cultured at 28°C for 24 hours, and the strain was detected by the Biolog Gram-negative plate The utilization rate of carbon source was compared with the Biolog identification database GN 4.01.
[0028] The results showed that the strain could utilize lactose and melibiose, and the strain was identified as Burkholderia cepacia, with a similarity index of 0.65.
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com