Method for amplifying complete sequence of mitochondrial genome of Macrobrachium nipponense
A technology of mitochondrial genome and mitochondrial gene, which is applied in the field of full-sequence amplification of the mitochondrial genome of Macrobrachium japonicus, and the field of full-sequence amplification of the mitochondrial genome, which can solve the problem of low yield of extracted mitochondrial genomic DNA, difficulty in repeating mitochondrial DNA in experiments, long operation time, etc. problems, to achieve the effect of not easy to degrade, the requirements of experimental conditions are not strict, and the operation time is short
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
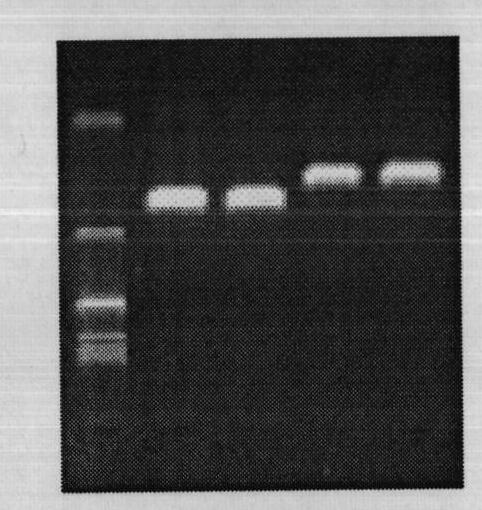
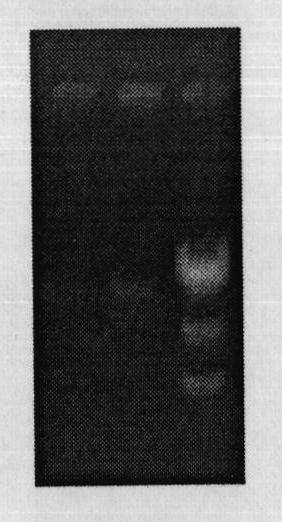
[0046] Experiment 1. Extraction of the mitochondrial genome of Macrobrachium japonicus by high-salt precipitation method
[0047] Use liquid nitrogen to grind fresh giant river prawn (Macrobrachium nipponense) muscles collected in Shanghai, China to powder, transfer 50 mg to a 1.5 mL centrifuge tube and add solution I (Tris-HCl 10 mmol / L, NaCl 10 mmol / L, MgCl 2 5mmol / L pH 7.5) 500μl, mix well, 2000r / min, 10min, discard the supernatant. Add solution II (Tris-HCl 10mmol / L, NaCl 400mmol / L, EDTA 2mmol / L pH 8.0) 500μl to the precipitation, mix well, 3500r / min, 10min, discard the supernatant. Precipitation plus solution II 100 μl, after mixing, add 5 μl of 20 mg / mL proteinase K and 20 μl of 10% SDS, and mix quickly. 55°C water bath for 1.5h. Add 50 μl saturated sodium acetate, mix well, and shake gently for 15S. Centrifuge at 15000r / min, 4°C for 15min, and take the supernatant into an Eppendrof tube. Add 50 μl saturated sodium acetate again, mix well, and shake gently for 15S. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com