Protein capable of promoting chloroplast development and coding gene and application thereof
A technology that encodes genes and proteins, which can be used in applications, genetic engineering, plant genetic improvement, etc., can solve few problems, and achieve the effects of increasing crop yield, improving photosynthetic capacity, and high economic benefits
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Embodiment 1, phenotype and genetic analysis of rice white stripe mutant ws
[0058] 1. Phenotype analysis of rice white stripe mutant ws
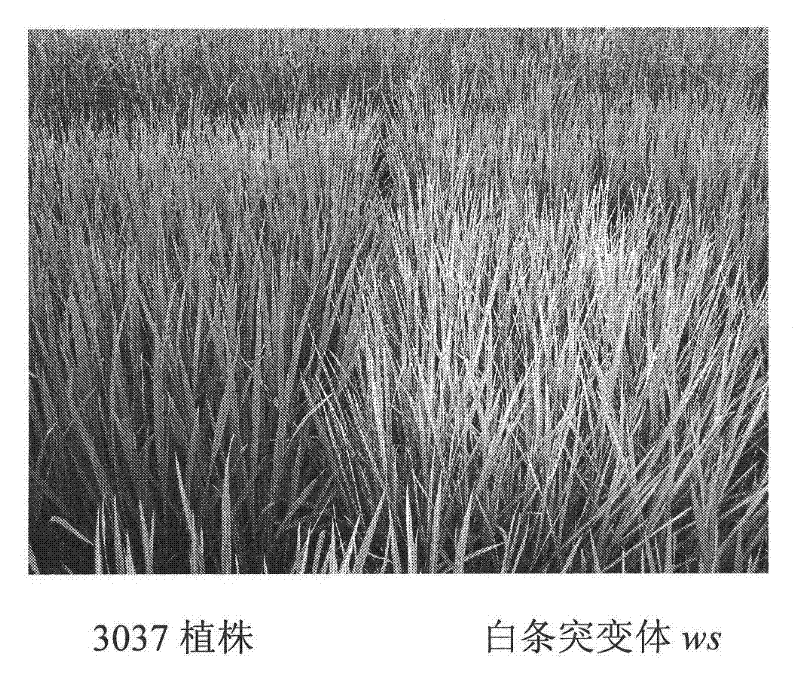
[0059] The rice white stripe mutant ws is a natural mutant of Zhongxian 3037 (WT, purchased from the Agricultural College of Yangzhou University) found in the field by our laboratory. Compared with the control 3037, the phenotype of the white-striped mutant was that white stripes were distributed on the leaves along the veins from the seedling stage. As the plant grew, the white stripes gradually increased on the leaves, and the most serious phenotype was the second leaf. The phenotype of the sword leaf is somewhat relaxed. At the booting stage, white stripes were distributed on most of the second leaves of the severe phenotype plants. The mutant phenotype does not affect the normal growth and development of rice under normal cultivation conditions ( figure 1 ). The analysis of the chlorophyll content of the leaves of the plant...
Embodiment 2
[0066] Example 2, the acquisition of ribonucleotide reductase small subunit WS and its coding gene WS
[0067] 1. Map-based cloning of WS genome genes
[0068] In order to clone the WS gene, we will use the homozygous white stripe mutant ws to cross with Nipponbare, and the obtained F 1 F 2 Population, among which 180 F2 recessive individuals (F2 generation individuals with white stripe phenotype) were initially mapped for WS gene. Using STS (Sequence-Tagged Site) molecular markers and using PCR method, we found that the STS markers S1, S2, S3 and S4 on chromosome 6 had obvious linkage with the mutation site. Most of the exchanged plants between the mutation site and S2 were also exchanged between the mutation site and S1, and most of the exchanged plants between the mutation site and S3 were included in the mutation site and S4 in exchange among individual plants. At the same time, the exchanged individual plants between the mutation site and S3 were different from those ...
Embodiment 3
[0076] Example 3, Complementation Experiment of White Stripe Mutant WS Phenotype
[0077] 1. Construction of complementary vector pCWS and complementary control vector pCWSC
[0078] BAC OsJNBb0055C04 (purchased from Shanghai National Gene Research Center, Chinese Academy of Sciences, No. OsJNBb0055C04) was digested with EcoR I to obtain 3363 bases upstream of the start codon ATG of WS and 3510 bases after the stop codon TGA. The DNA fragment (7893bp) of the full-length sequence was cloned between the EcoR I recognition sites of pCAMBIA1300 (DingGuo, MCV033), and the complementary expression vector pCWS was constructed. The constructed complementary vector pCWS was digested with PstI to remove part of the coding region of the WS gene and the 5' promoter region of the gene, and retain the part of the coding region and the 3' regulatory region at the 3' end, thus constructing the complementary control vector pCWSC( Figure 4 Middle C).
[0079] 2. Obtaining and phenotypic ident...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com