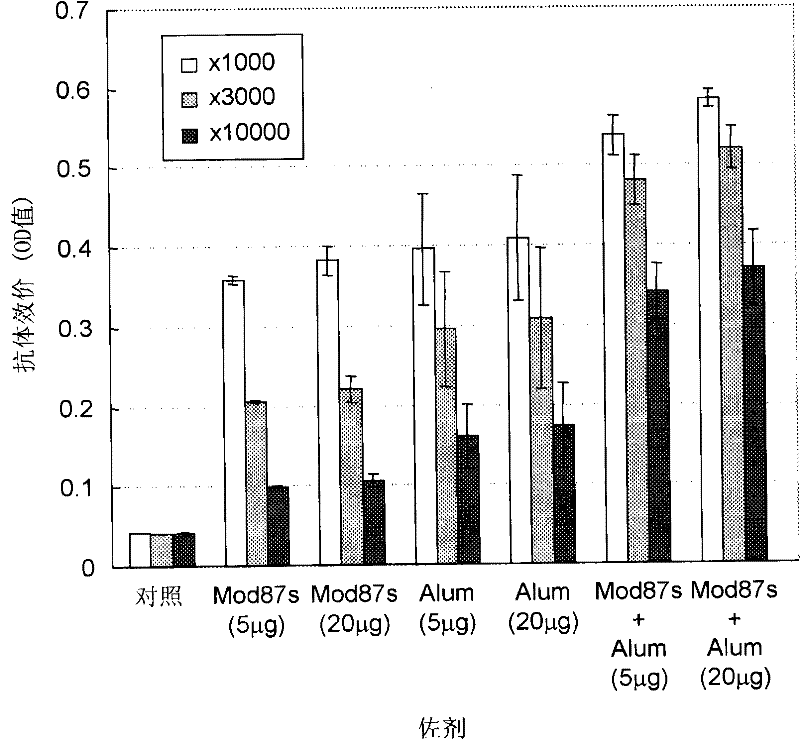
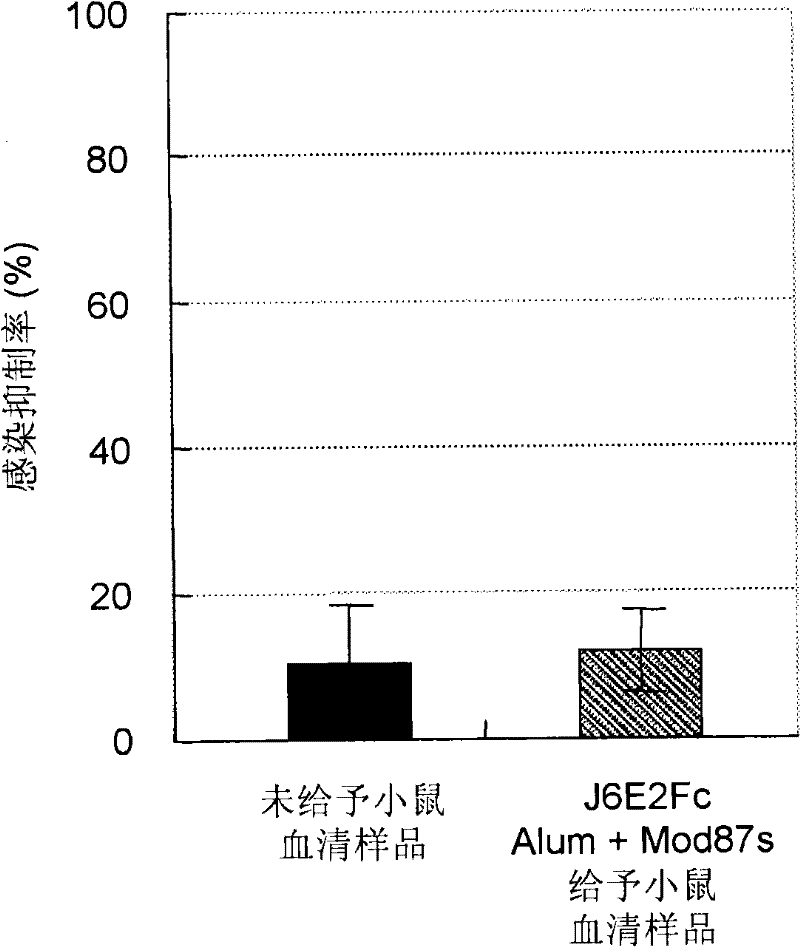
Hepatitis C virus vaccine composition
A technology of hepatitis C virus and vaccine composition, which is applied in the direction of virus, virus peptide, drug combination, etc., can solve the problem of HCV infection inhibitory activity of undisclosed antibodies, and achieve the effect of preventing virus infection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0126] Preparation of J6 / JFH1-HCV particles
[0127] The cDNA (genomic cDNA) obtained by reverse transcription of the entire genomic RNA region of the hepatitis C virus (HCV) JFH1 strain (genotype 2a) isolated from patients with fulminant hepatitis was cloned into the downstream of the T7 RNA promoter sequence of the pUC19 plasmid, and the obtained The plasmid DNA pJFH1 (Wakita, T. et al., Nat.Med., 11 (2005) 791-796 pages and International Publication WO 2004 / 104198) was digested with EcoR I, and further partially digested with Bcl I to prepare the excised A plasmid DNA fragment of a fragment (about 2840 bp) from the EcoR I site to the original BclI site was purified.
[0128] On the other hand, genomic cDNA from HCV J6CF strain (GenBank accession number AF177036, Yanagi, M., et al., Virology 262:250-263 (1999)) was cloned into the downstream of the T7 RNA promoter sequence of pUC19 plasmid, and The obtained plasmid DNA, i.e. pJ6CF (International Publication WO 2006 / 022422),...
Embodiment 2
[0135] Purification of J6 / JFH1-HCV particles
[0136] The J6 / JFH1-HCV particles produced in Example 1 were purified by the following 3 steps.
[0137] 1) Concentration and diafiltration
[0138] Using a hollow fiber cartridge (Hollow Fiber Cartridge) (GE Healthcare company: 500kDa cut-off, model UFP-500-C-8A, hereinafter referred to as "hollow fiber"), the culture supernatant containing HCV particles obtained in Example 1 above was concentrated to 60 times.
[0139] 2) Density gradient ultracentrifugation
[0140] Add 3 mL of cooled TNE buffer (10 mM Tris-HCl (pH7.4), 150 mM NaCl, 1 mM EDTA) containing 60% sucrose to an Ultra-clear 25×89 mm centrifuge tube (Beckman Coulter Company: catalog number 344058), covering 7 mL of TNE buffer containing 20% sucrose. Another 25 mL sample was overlaid on TNE buffer containing 20% sucrose. Using SW-28 (Beckman Coulter Co.), ultracentrifugation was performed at 28,000 rpm at 4° C. for 4 hours.
[0141] A 25G injection needle (Teru...
Embodiment 3
[0145] Inactivation of HCV particles
[0146] The concentrated hepatitis C virus obtained in steps 1) to 3) of Example 2 was inactivated by ultraviolet irradiation. GL-15 manufactured by Toshiba Corporation was used as a line source of ultraviolet rays. Specifically, the purified infection titers were 1 × 10 6 The ffu / mL solution containing hepatitis C virus particles (JFH1 strain) was placed in a silicon-coated polyethylene microcentrifuge tube (manufactured by Assist), and placed in a place where the irradiation intensity of ultraviolet light was 20 mW / cm 2 distance, irradiate UV-C for 5 minutes.
[0147] After treatment, the hepatitis C virus particles were serially diluted 50 times, 250 times, 1250 times, 6250 times, 31250 times, 156250 times and 781250 times in Dulbecco's modified Eagle medium (DMEM).
[0148] The day before, press 1×10 4 Cells / well were placed in a 96-well poly-L-lysine coated plate (manufactured by Corning; Corning 96 Well Clear Flat Bottom Poly-L-l...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com