High-temperature resistance arabinfuranosidease Abf51B8, as well as gene and application thereof
A furanosidase and high temperature-resistant technology, applied in the field of genetic engineering, can solve the problems of poor thermal stability and inability to meet the requirements
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Example 1 Cloning of Alicyclobacillus sp.B8 Arabinofuranosidase Encoding Gene abf51B8
[0042] Extract the genomic DNA of Alicyclobacillus sp.B8:
[0043] Centrifuge the bacteria cultured in the liquid for 3 days, put them into a mortar, add 2mL extract, grind for 5min, then put the grinding solution in a 50mL centrifuge tube, lyse in a water bath at 65°C for 20min, mix well every 10min, and Centrifuge at 10,000 rpm for 5 min at 4°C. The supernatant was extracted in phenol / chloroform to remove impurities, and then an equal volume of isopropanol was added to the supernatant. After standing at room temperature for 5 minutes, centrifuge at 10,000 rpm for 10 minutes at 4°C. The supernatant was discarded, the precipitate was washed twice with 70% ethanol, dried in vacuo, dissolved by adding an appropriate amount of TE, and stored at -20°C for later use.
[0044] The degenerate primers P1, P2 were designed and synthesized according to the conserved (GNEMDG and DEWNVW) seque...
Embodiment 2
[0052] The preparation of embodiment 2 recombinant arabinofuranosidase
[0053] The expression vector pPET-30a is subjected to double digestion (Nde I+Not I), and the gene abf51B8 encoding arabinofuranosidase is double-digested (Nde I+Not I) at the same time, and the gene fragment encoding mature arabinofuranosidase is cut out Ligated with the expression vector pPET-30a, the recombinant plasmid pPET-abf51B8 containing the arabinofuranosidase gene abf51B8 was obtained and transformed into Escherichia coli BL21(DE3) to obtain the recombinant Pichia pastoris strain BL21 / abf51B8.
[0054] The BL21 strain containing the recombinant plasmid was inoculated in 300mL LB culture medium, shaken at 227rpm at 37°C for 2h, and then induced with a final concentration of 0.6mM IPTG at 30°C for 4h. The expression level of recombinant arabinofuranosidase was 5.6U / mL. The results of SDS-PAGE showed that the recombinant arabinofuranosidase was expressed in Escherichia coli. The specific activit...
Embodiment 3
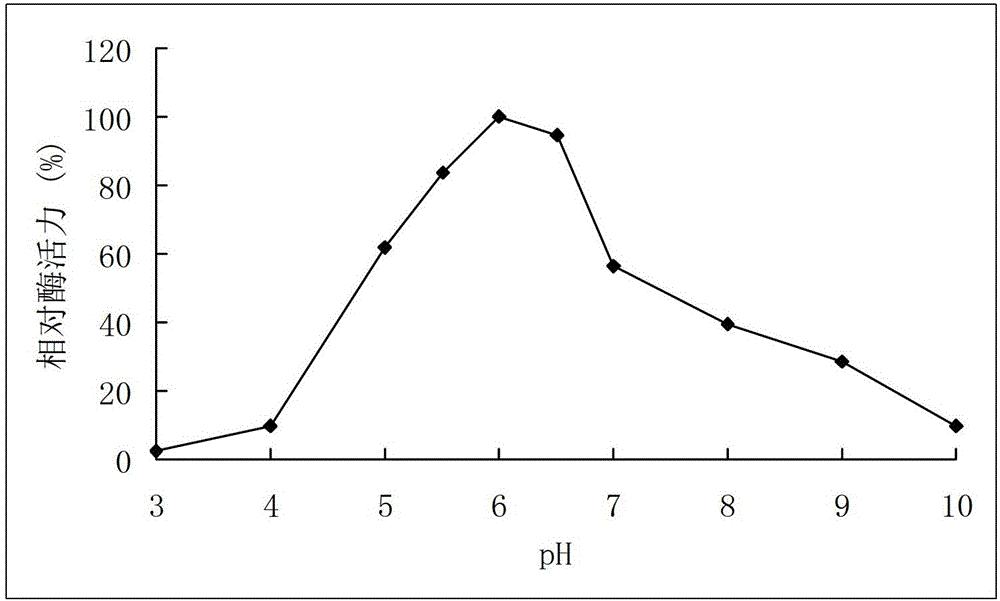
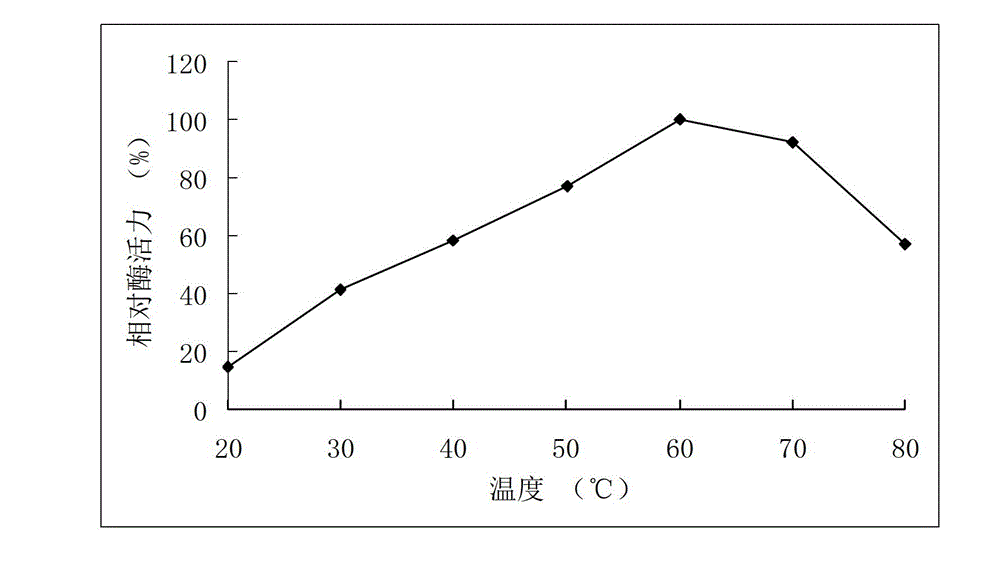
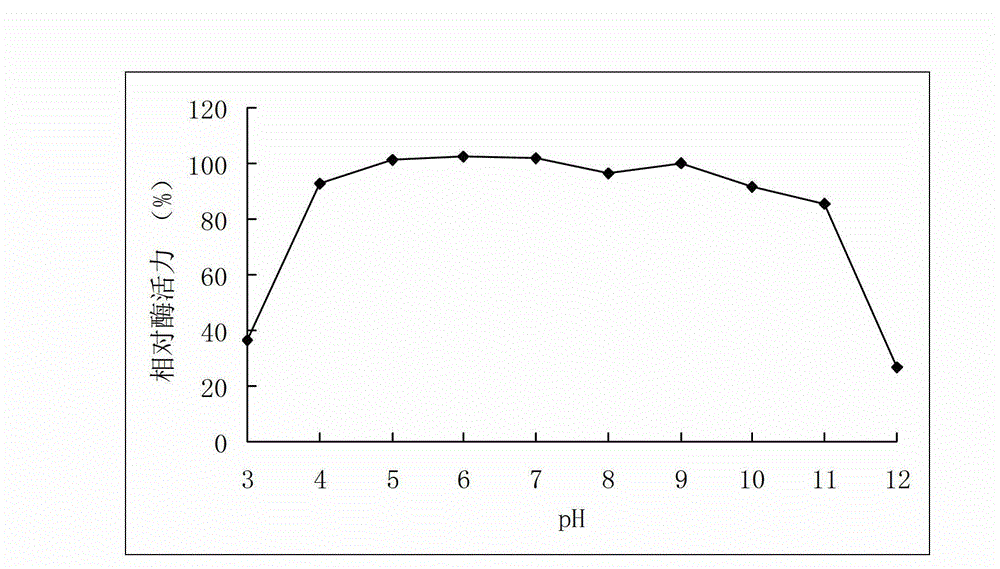
[0055] Example 3 Activity analysis of recombinant arabinofuranosidase
[0056] Determination of the activity of arabinofuranosidase: the amount of the product p-nitrophenol generated by the enzymatic hydrolysis of the substrate pNPAf was measured at 405 nm. Reaction steps: Mix 250μL 2mM pNPAf substrate with 150μL buffer, add 100μL appropriately diluted enzyme solution, react at 40°C for 10min, add 1.5mL 1M Na 2 CO 3 The reaction was terminated, and the OD value was measured at 405 nm.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Theoretical molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com