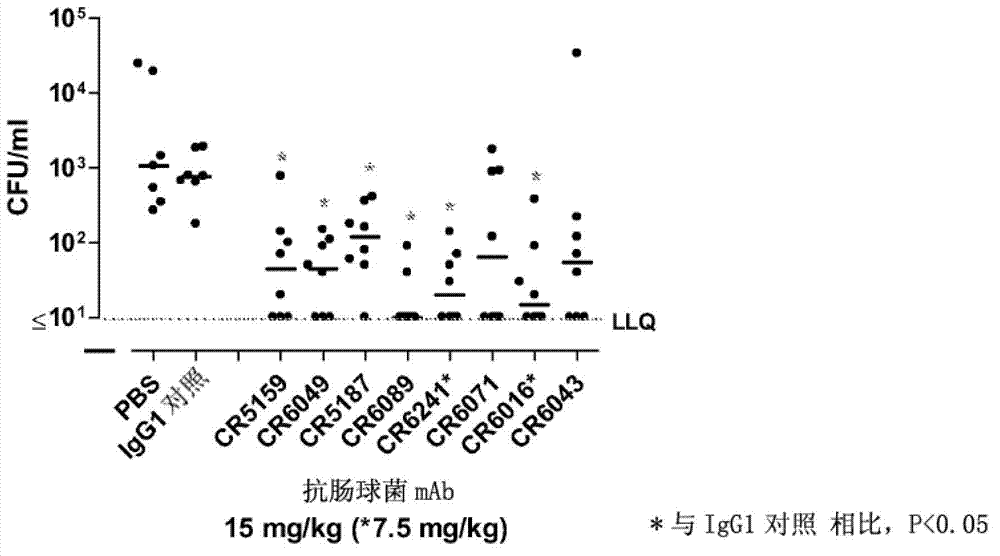
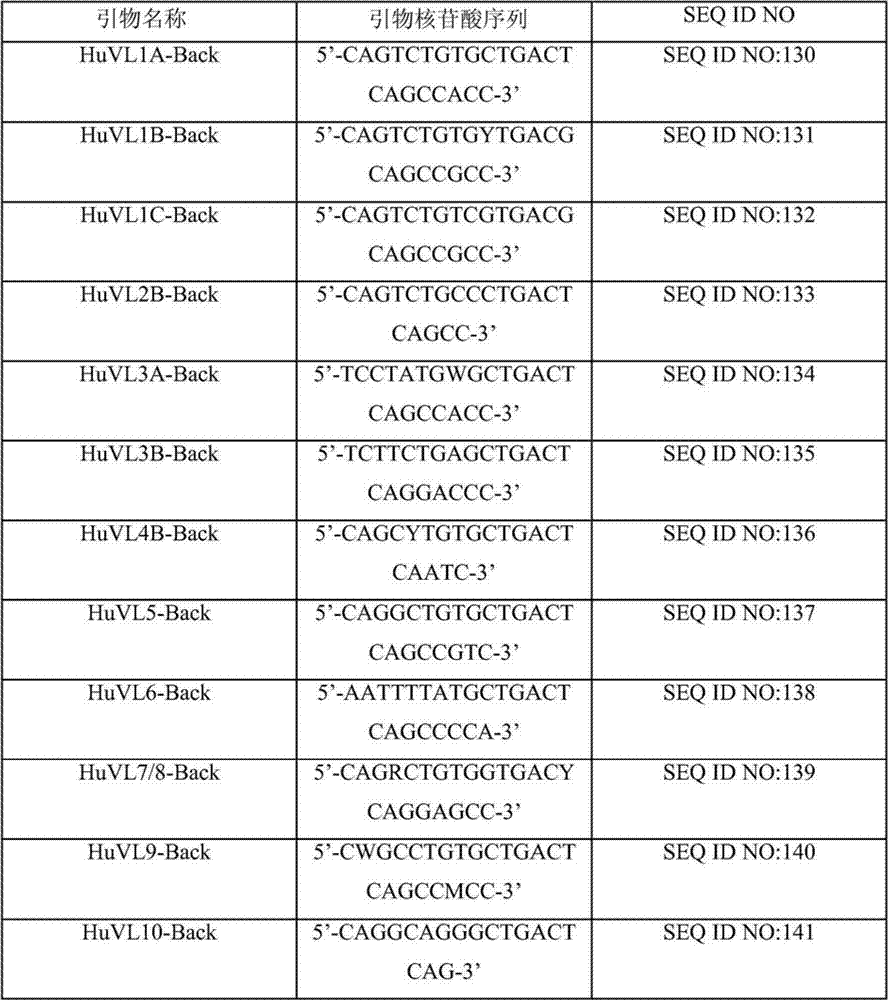
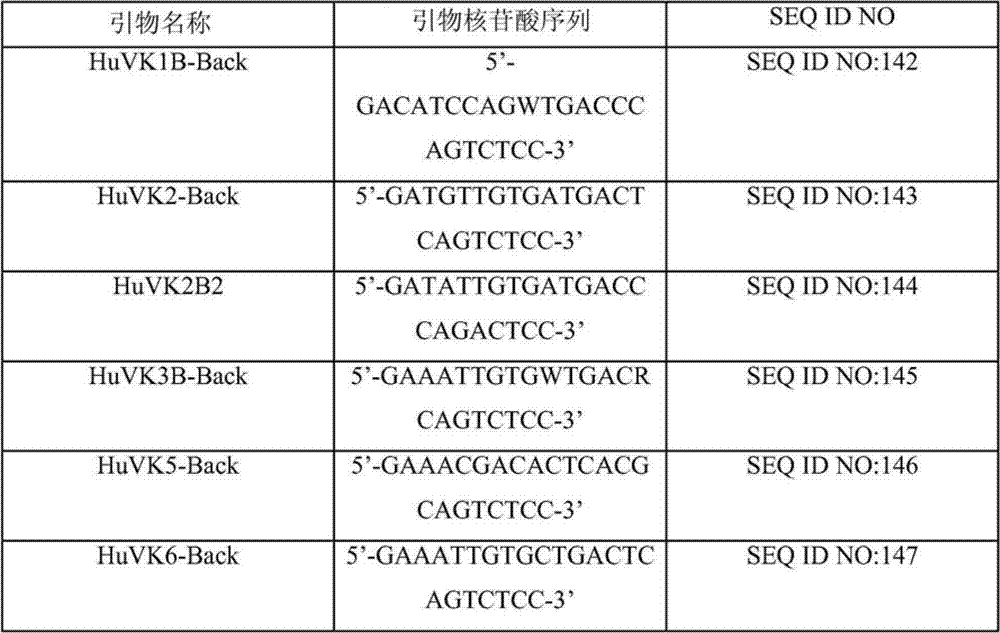
Human binding molecules having killing activity against enterococci and staphylococcus aureus and uses thereof
A staphylococcus and enterococcus technology, applied in the fields of application, bacterial antigen components, bacteria, etc., can solve the problems of unpublished human antibodies, unpublished sequences, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0134] Construction of scFv Phage Display Libraries Using RNA Extracted from Donors for Screening for Opsonization Activity
[0135]Blood samples were drawn from donors aged 25-50 years who reported recent Gram-positive bacterial infection as well as healthy adults. Peripheral blood leukocytes were separated by centrifugation, and serum was stored and frozen at -80°C. Donor sera were screened for killing activity using the opsonophagocytic killing assay (Huebner et al., 1999) and compared to normal rabbit sera. Sera from donors with higher than normal phagocytic activity were selected for the generation of phage display libraries. Total RNA was prepared from peripheral blood leukocytes of these donors using organic phase separation followed by ethanol precipitation. The resulting RNA was dissolved in RNase-free water and its concentration determined by OD260nm detection. After that, the RNA was dissolved to a concentration of 100 ng / µl. Next, 1 μg of RNA was converted into...
Embodiment 2
[0139] Construction of scFv phage display libraries using RNA extracted from memory B cells
[0140] Peripheral blood was collected from normal healthy donors, recovering donors, or vaccinated donors by venipuncture using EDTA anticoagulated tubes. Blood samples (45ml) were diluted twice with PBS and 30ml aliquots placed under 10ml Ficoll-Hypaque (Pharmacia) and centrifuged at 900 xg for 20 minutes at room temperature without pause. The supernatant was carefully removed to just above the fraction containing lymphocytes and platelets. Next, carefully remove this layer (about 10 ml) and transfer to a new 50 ml tube, wash 3 times with 40 ml PBS, and centrifuge at 400 xg for 10 minutes at room temperature to remove platelets. The resulting lymphocyte-containing pellet was resuspended in RPMI medium containing 2% PBS, and the cell number was determined by cell counting. Use CD24, CD27, and surface IgM as markers to separate switched and IgM memory B cells for approximately 1X10 ...
Embodiment 3
[0142]Selection of phages carrying single-chain Fv fragments that specifically bind enterococci
[0143] Using antibody phage display libraries, common phage display techniques, and Techniques for selection of antibody fragments are primarily described in US Patent No. 6,265,150 and in WO 98 / 15833 (both incorporated herein by reference). The antibody phage libraries used were the screened donor library prepared as described in Example 1 and the IgM memory library prepared as described in Example 2. The methods and helper phages described in WO 02 / 103012 (incorporated herein by reference) are used in the present invention. To identify phage antibodies that recognize enterococci, phage selection experiments were performed using live bacteria in suspension or immobilized bacteria in immunotubes. The strains used are described in Table 8. All phage antibodies were isolated from selections in which, in at least one step, E. faecalis 12030 in suspension was used. Antibodies des...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com