Applications of ARID1A gene and protein product encoded by the same
A technology of encoding protein and protein products, which is applied in the field of tumor diagnosis and treatment, can solve the problems of liver cancer occurrence, metastasis, and unclear genetic events of liver cancer, and achieve the effect of inhibiting proliferation and metastasis and accurate diagnosis
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 110
[0045] Example 110 Exome Capture Sequencing of Liver Cancer Patients
[0046] 1. Tissue sample collection and DNA extraction
[0047] Collect paracancerous tissue, cancer tissue, and portal vein tumor thrombus tissue samples from 10 patients with liver cancer (30 tissue samples in total), and use the tissue DNA extraction kit from QIAGEN, Germany, to perform tissue DNA extraction according to the experimental procedures in the product manual . The quality of the extracted DNA was verified. Except for the DNA degradation of the portal vein tumor thrombus tissue in one patient, the DNA quality of the other 29 tissue samples met the requirements.
[0048] 2. Exome Capture
[0049] Genomic DNA was randomly broken into fragments of about 500bp, and then adapters were connected to both ends of the DNA fragments. The DNA fragments that passed the PCR library check were hybridized with the NimbleGen 2.1M Human Exome Array chip. After removing the background DNA not bound to the ch...
Embodiment 2
[0063] The Sanger method sequencing of embodiment 2PCR product
[0064] 1. PCR amplification of specific DNA fragments in the coding region of the ARID1A gene
[0065] The primer sequences are:
[0066] Forward primer: TGCGTGTCCTTTGTTATATTGG (SEQ ID NO: 1);
[0067] Reverse primer: TCAGATCAGTCACCTTTTCCTCA (SEQ ID NO: 2).
[0068] High-fidelity hot-start DNA polymerase Hotstar Taq is used.
[0069] The PCR reaction system is: forward primer, reverse primer, 10X PCR buffer, Mgcl 2, dNTP, Hotstar Taq DNA polymerase, and DNA template, the volumes of each component are 0.5, 0.5, 5, 1, 0.5, 0.1, 1 μl, and finally add sterilized deionized water to make the reaction system 50 μl.
[0070] The PCR reaction conditions were: denaturation at 95°C for 5 minutes, followed by 30 cycles of denaturation at 95°C for 30 seconds, annealing at 55°C for 30 seconds, and extension at 72°C for 30 seconds in each cycle; PCR amplification products were detected by electrophoresis and PCR products we...
Embodiment 3
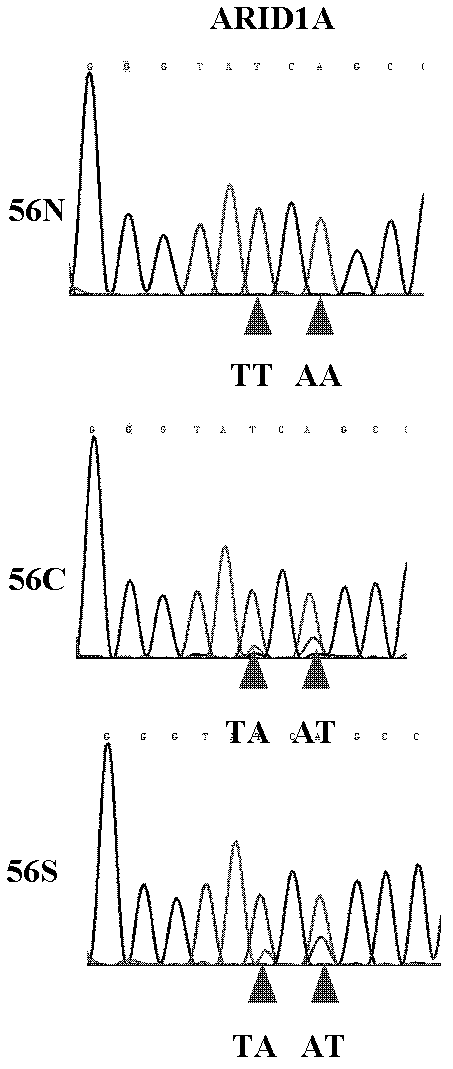
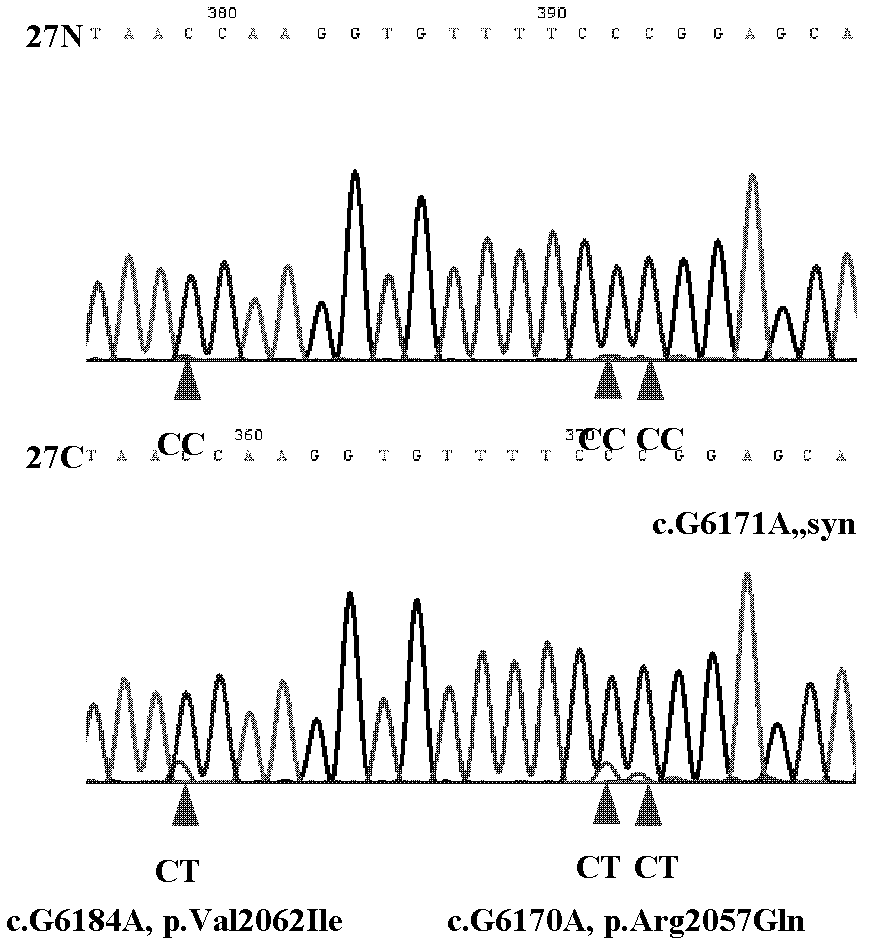
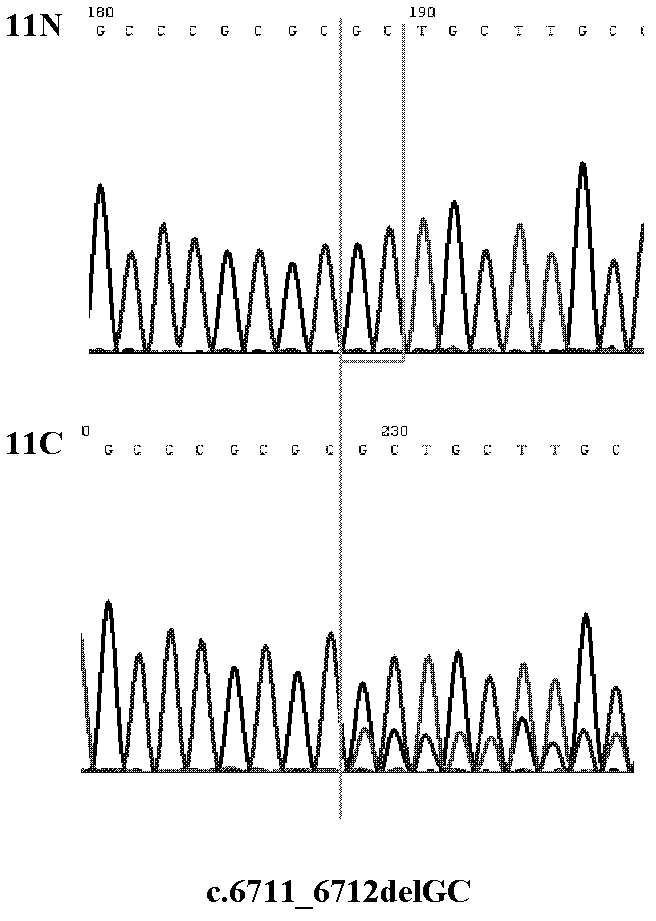
[0075] Example 3 Sequencing Analysis of ARID1A Gene Coding Sequence in Liver Cancer Patient Tissue by Sanger Method
[0076] 1. Tissue Sample Collection
[0077] The tissue samples of 100 pairs of liver cancer patients were obtained from surgically excised cancer tissue and paracancerous tissue samples of liver cancer patients in hospitals in Guilin, Guangxi, Wuxi, Jiangsu, and Suzhou, Jiangsu. All tissue samples were frozen and stored at -80°C. The human tissue manipulations involved were approved by the Ethics Committee of the National Human Genome Southern Research Center.
[0078] 2. DNA extraction
[0079] Tissue DNA extraction kits from QIAGEN, Germany were used to extract tissue DNA according to the experimental procedures in the product manual.
[0080] 3. PCR amplification of the nucleic acid sequence encoded by the ARID1A gene
[0081] The ARID1A gene consists of 20 exons (expressed region, Exon), and the encoded nucleic acid sequence (CCDS285.1) is derived from t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com