Cordyceps Chinese Hirsutella nucleoside diphosphokinase, coding gene and application thereof
A technology of Mortierella diphosphate nucleoside kinase and Cordyceps sinensis, applied in application, genetic engineering, plant gene improvement, etc., can solve the problems of rare gene and protein research, achieve great application prospects, expand production, and high expression Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Example 1: Cultivation of "Bailing" production fungus Cordyceps sinensis
[0035] Source of the strain: Firstly, natural Cordyceps sinensis was collected from Qinghai, and brought back to Hangzhou for isolation and screening, and the L0106 strain was obtained, and the strain was identified as Hirsutella Sinensis, which was preserved in a typical culture in China Depository Center, the preservation number is CCTCC No: M 2011278, which has been disclosed in the previously applied patent CN102373190A.
[0036] Inoculate the strain on the slant, and the medium formula (this is the liquid formula before solidification, prepared according to the following proportions before making the slant) is glucose 2.0% (w / v, 1% means that 100mL medium contains 1g , the same below), corn flour 1.0%, potato juice 0.5%, dextrin 0.5%, yeast powder 0.5%, bran 1.0%, silkworm chrysalis powder 2.0%, peptone 1.0%, magnesium sulfate 0.05%, potassium dihydrogen phosphate 0.05% , agar powder 1.0%, ...
Embodiment 2
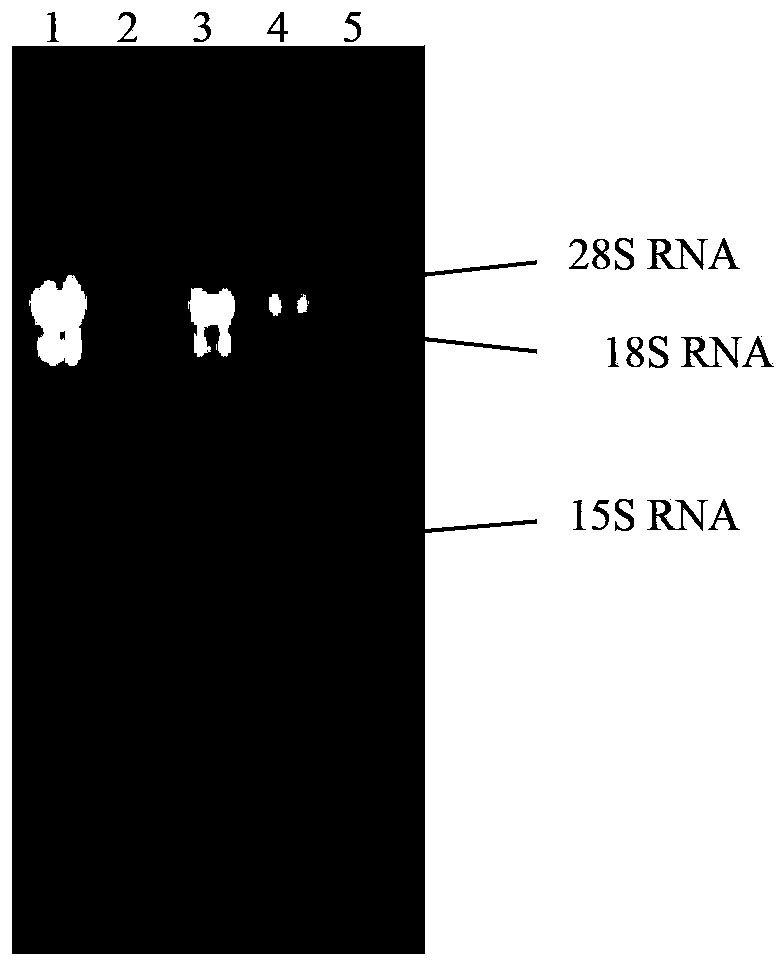
[0037] Example 2: Extraction of total RNA of "Bailing" production fungus Cordyceps sinensis
[0038] The total RNA was extracted with TRIzol reagent, and the steps were as follows: 1) Grinding with liquid nitrogen: Take 1 g of fresh bacteria and put it into a mortar, add liquid nitrogen repeatedly to grind until powdery, and divide into pre-cooled 1.5mL centrifuge tubes, Add 1mL TRIzol reagent, mix well, and let stand on ice for 5min to completely separate the nucleic acid-protein complex. 2) RNA isolation: add 0.2mL chloroform, shake vigorously for 15s, let stand on ice for 2~3min, centrifuge at 12000rpm at 4°C for 15min, separate the layers, and take the upper aqueous phase, about 600μL. 3) RNA precipitation: add 500 μL of isopropanol, let stand on ice for 10 minutes, centrifuge at 12000 rpm at 4°C for 10 minutes, and discard the supernatant. 4) RNA washing: add 1 mL of 75% (v / v) ethanol, suspend the pellet, let stand on ice for 10 min, centrifuge at 4°C, 7500 rpm for 15 mi...
Embodiment 3
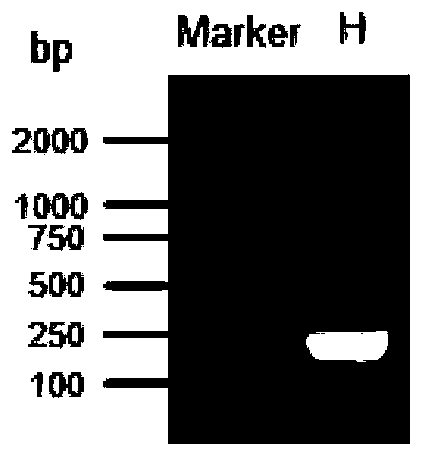
[0039] Example 3: Sequencing of the RNA sample of "Bailing" production fungus Cordyceps sinensis
[0040] After extracting the total RNA from the sample, the mRNA was enriched with Oligo(dT) magnetic beads. Add fragmentation buffer to break mRNA into short fragments (200~700bp), use mRNA as a template, use six base random primers (random hexamers) to synthesize the first cDNA strand, then synthesize the second cDNA strand, and then pass through QiaQuick PCR After the kit is purified and eluted with EB buffer, end repair is performed, polyA is added and sequencing adapters are connected, and then agarose gel electrophoresis is used for fragment size selection, and finally PCR amplification is performed. The built sequencing library is carried out with Illumina GA IIx sequencing. The original image data obtained by sequencing is converted into sequence data through base calling, that is, raw data or raw reads. The reads containing only the adapter sequence in the original sequ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com