Recognition probe, detection method and application of cancer cells
A technology of tumor cells and detection methods, which is applied in biochemical equipment and methods, measurement/inspection of microorganisms, DNA/RNA fragments, etc., can solve the problems of low detection sensitivity, achieve high detection sensitivity, high detection efficiency, and chemical stability good sex effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Human lung cancer cells were used as tumor cells to be tested, and a recognition probe (recognition probe 1) whose sequence was SEQ ID No.2 was designed based on the nucleic acid aptamer S11e (SEQ ID No.1) of human lung cancer A549 cells.
[0055] The length of S11e is 45bp, and a 32bp extended sequence is added to the 5' end of S11e to form probe 1.
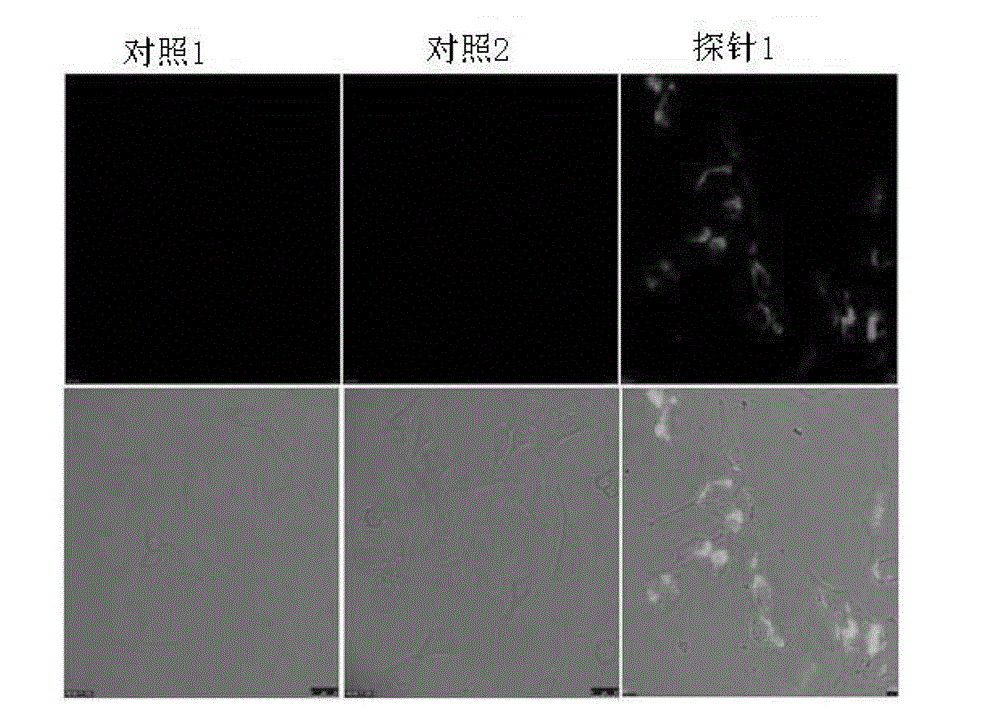
[0056] Specific binding experiment of probe 1 to human lung cancer A549 cells
[0057] Specific steps are as follows:
[0058] Label the 5' end of probe 1 with fluorescein FAM to form FAM-probe 1;
[0059] Human lung adenocarcinoma A549 cells were cultured in laser confocal dishes;
[0060] Wash human lung adenocarcinoma A549 cells 3 times with phosphate buffered solution PBS;
[0061] Dilute FAM-probe 1 with phosphate buffer;
[0062] Mix the phosphate buffer solution of FAM-probe 1 with human lung cancer A549 cells, and incubate at room temperature for 20 minutes; use the lung cancer A549 cells without FAM-probe 1...
Embodiment 2
[0084] Human lung cancer cells were used as tumor cells to be tested, and a recognition probe (recognition probe 2) whose sequence was SEQ ID No.5 was designed based on the nucleic acid aptamer S11e (SEQ ID No.1) of human lung cancer A549 cells.
[0085] In order to eliminate primer-dimer interference during real-time fluorescent quantitative PCR, the sequence of the recognition probe was increased. The length of S11e was 45 bp, and a 55 bp extended sequence was added to the 5' end of S11e to form probe 2.
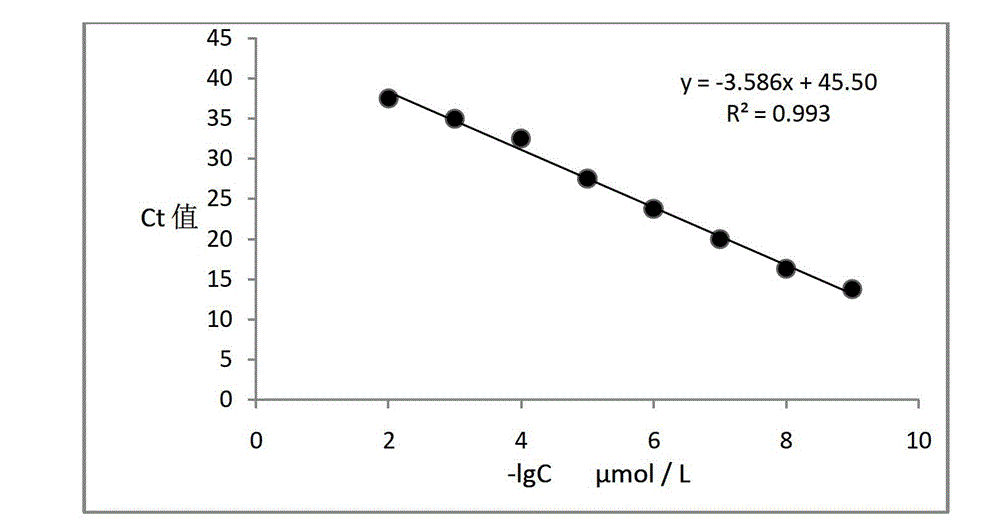
[0086] PCR amplification experiment using probe 2 as template
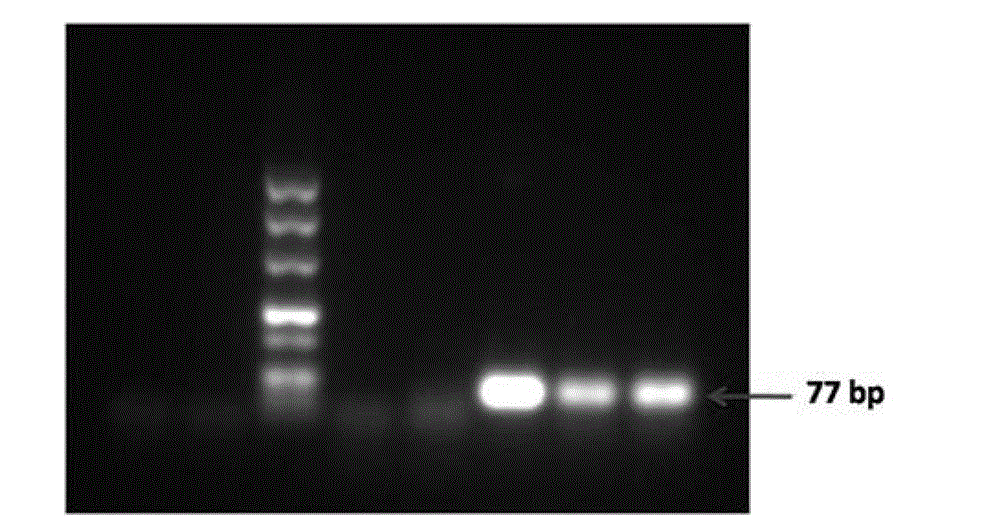
[0087] According to the sequence of probe 2, an upstream primer whose sequence is shown in SEQ ID No.6 and a downstream primer shown in SEQ ID No.7 were designed. The PCR reaction system is shown in Table 1, and the PCR reaction conditions are shown in Table 2.
[0088] After PCR, the PCR products were electrophoresed with 2% agarose gel, and the results were as follows: Figure 4 As shown, the 100bp band i...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com