Chip-based 5-hydroxymethylated cytosine detection method with high flux and high sensitivity
A technology for hydroxymethylated cytosine and sensitive detection is applied in the field of high-throughput and highly sensitive detection of 5-hydroxymethylated cytosine, and can solve the problems of false positive detection results, high detection cost, inaccurate detection results, and the like, To achieve the effect of high specificity, high sensitivity detection, high sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
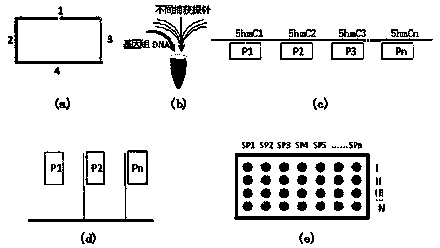
Image
Examples
Embodiment 1
[0034] To achieve the detection of the hydroxymethylated cytosine (hmC) status of the first C site (194-59366) of the 68 CCGG sequences of the BDNF gene (NT_039207.8) in the spinal cord of mice with chronic inflammatory pain.
[0035] (1) Design specific amplification primers with acrylamide modification at the 5'end for the first C sites of the above 68 CCGG sequences, dilute them to 20uM and fix them on a glass slide treated with acrylamide to form a microarray chip. Two open-loop detection probes are designed for each C site of the above 68 CCGG sequences, namely, open-loop detection probe I and open-loop detection probe II (the open-loop probe I is used to detect the non-hmC state, Open-loop probe II is used to detect hmC status. The main sequence of the open-loop detection probe is shown in Table 1.
[0036] (2) Treat 1ug of mouse spinal cord DNA with T4-BGT, that is, treat the genomic DNA of the mouse to be tested with β-glucosyltransferase, and sugar all 5-hydroxymethylated ...
Embodiment 2
[0041] Realize the detection of the hydroxymethylated cytosine (hmC) status at the first C site of 33 CGCG sequences in the plasma DNA of patients with chronic rheumatoid pain in the BDNF gene (AC_000143.1).
[0042] (1) Design specific amplification primers with acrylamide modification at the 5'end for the first C sites of the 33 CGCG sequences, diluted to 20uM and fixed on the glass slide treated with acrylamide to form a microarray chip. Then, two open-loop detection probes are designed for each C site of the 33 CGCG sequences, namely, open-loop detection probe I and open-loop detection probe II (the open-loop probe I is used to detect non-hmC states). , Open-loop probe II is used to detect hmC status. The main sequence of the open-loop detection probe is shown in Table 1.
[0043] (2) Treat 1ug patient plasma DNA with T4-BGT, that is, treat the genomic DNA of the mouse to be tested with β-glucosyltransferase, and perform glycosylation of all 5-hydroxymethylated cytosines in the...
Embodiment 3
[0047] Achieve joint detection of 19 CCGG sequences (40945-59475) of the first C and COMT genes (NT_187012.1) of the first C and 10 CCGG sequences (771-15422) of the BDNF gene of the spinal cord DNA of mice with chronic neuropathic pain .
[0048] (1) For the 19 CCGG sequences (40945-59475) of the aforementioned BDNF gene, the first C and the first C of COMT gene (NT_187012.1) and 10 CCGG sequences (771-15422) were designed with acrylamide at the 5'end. The modified specific amplification primers were diluted to 20uM and fixed on the glass slide treated with acrylamide to form a microarray chip. Two open-loop detection probes are designed for each C site, namely open-loop detection probe I and open-loop detection probe II (where open-loop probe I is used to detect non-hmC states, and open-loop probe II Used to detect hmC status. The main sequence of the open loop detection probe is shown in Table 1.
[0049] (2) Treat 1ug of mouse spinal cord DNA with T4-BGT, that is, treat the g...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com