Short-chain dehydrogenase, coding gene, vector, engineering bacterium and application derived from Refsonia
A technology of short-chain dehydrogenase and Refsonia spp., applied in the field of short-chain dehydrogenase, can solve the problems of low activity of short-chain dehydrogenase/reductase, limitation of large-scale preparation and application, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Example 1: Short-chain dehydrogenase and coding gene derived from Refsonia
[0058] The total genomic DNA of the LeifsoniaxyliHS0904 strain was extracted with a nucleic acid extraction kit (AxygenCo.) (this strain has been protected as a new strain in the Chinese invention patent ZL201010523043.X and preserved in the Chinese Type Culture Collection Center, address: China, Wuhan, Wuhan University, preservation number CCTCCM2010241, preservation date: September 21, 2010), using the genomic DNA as a template, under the action of primer 1 (ATGGCNCARTAYGAYGTNGC) and primer 2 (YYAYTGNGCNGTRTAKCCYCC) PCR amplification.
[0059] Addition amount of each component of the PCR reaction system (total volume 50 μL): PfuDNA Polymerasemix 25 μL, cloning primer 1 and primer 2 each 3 μL (100 μM), genomic DNA 1 μL, nucleic acid-free water 18 μL. Biorad PCR instrument was used to amplify, and the PCR reaction conditions were: pre-denaturation at 95°C for 5min, then temperature cycle at 95...
Embodiment 2
[0060] Embodiment 2: Vector and recombinant genetically engineered bacteria
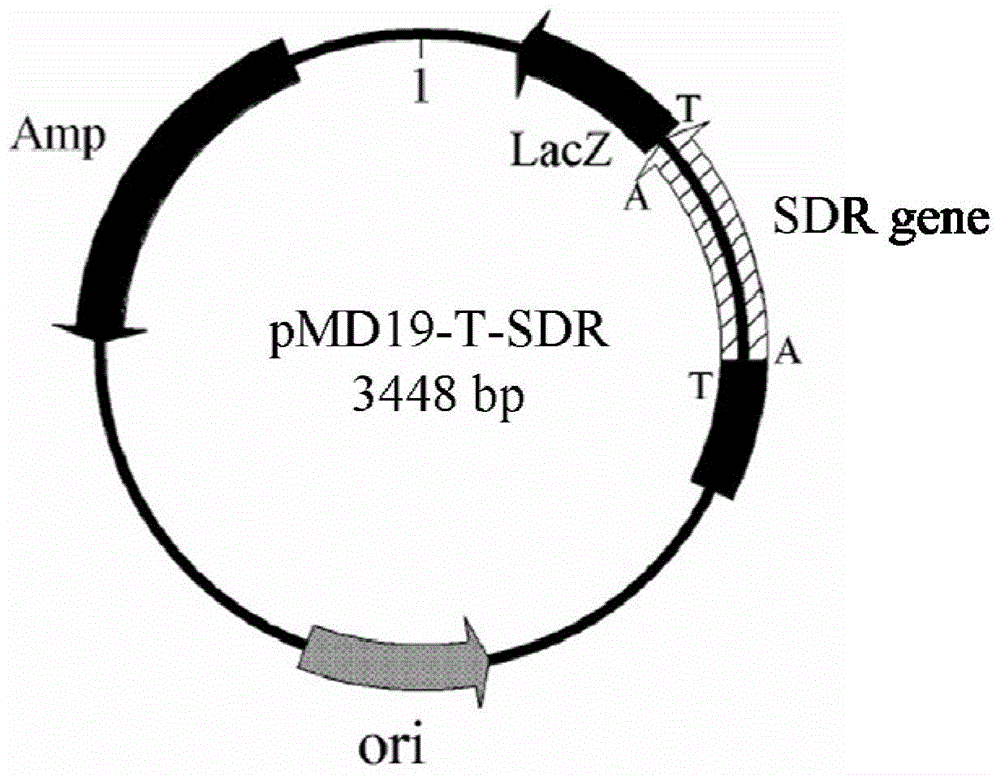
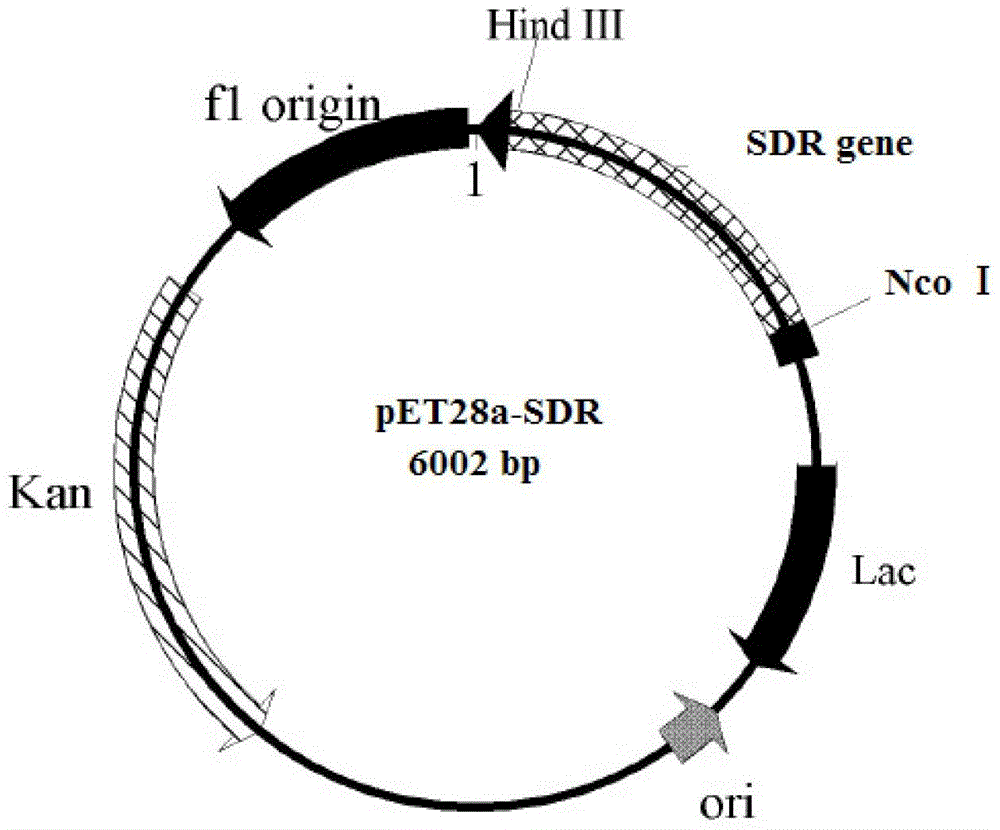
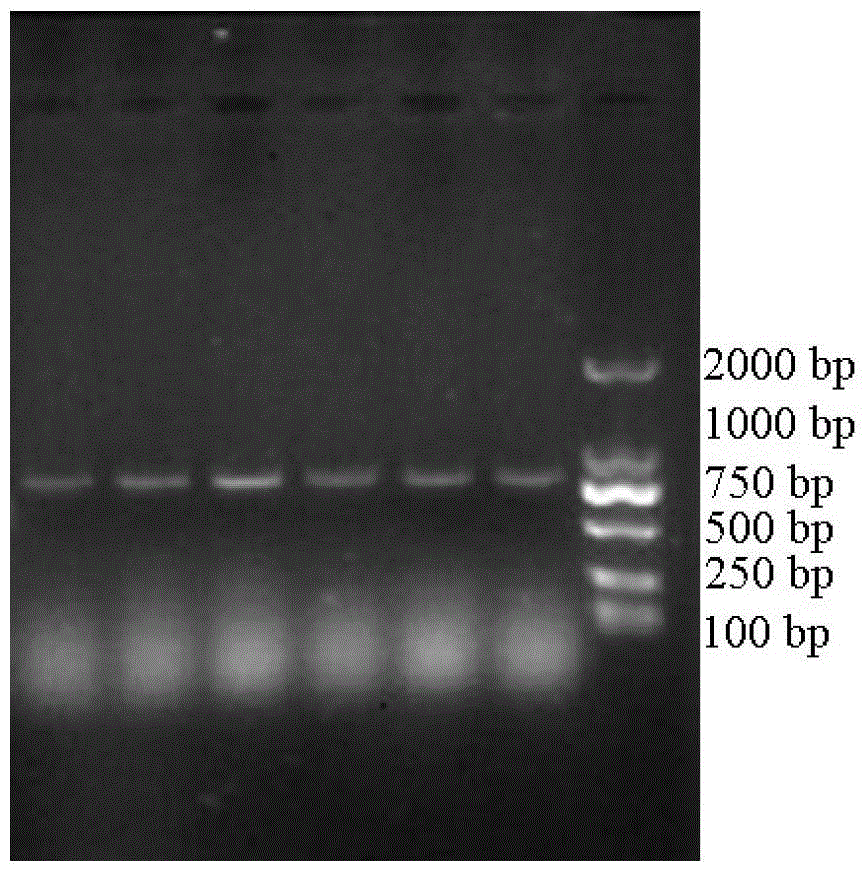
[0061] Expression primers 3 and 4 were designed according to the analysis results of Example 1, and NcoI and HindIII restriction enzyme sites were introduced into primers 3 and 4, respectively. In primer 3( CCATGG CGCAGTACGACGTGG, the underlined part is the NcoI restriction site, the italic part is the protected base) and primer 4 ( AAGCTT CTATTGTGCTGTGTAGCCTC, the underlined part is the HindIII restriction site, and the italic part is the protective base), under the triggering of high-fidelity Pyrobest DNA polymerase, a short-chain dehydrogenase gene fragment (SEQ ID NO.1) with a length of 756bp was obtained. After sequencing, use NcoI and HindIII restriction enzymes to treat the amplified fragment, and use T4 DNA ligase to connect the fragment with the expression vector pET28a(+) treated with the same restriction enzyme to construct the expression vector pET28a (+)-SDR, pET28a(+)-SDR recombi...
Embodiment 3
[0063] The recombinant Escherichia coli BL21 / pET28a(+)-SDR containing the expression recombinant plasmid pET28a(+)-SDR verified in Example 2 was used at 37°C and 200rpm in LB liquid medium containing 50 μg / mL kanamycin Shake culture for 12 hours, then inoculate into fresh LB liquid medium containing 50 μg / mL kanamycin with a volume concentration of 1% inoculum, and cultivate at 37°C and 200 rpm to a cell concentration of OD 600 0.6-0.9, then add isopropyl-B-D-thiogalactopyranoside (IPTG) with a final concentration of 0.1mM to the culture medium, place at 30°C, 200rpm for 10h, centrifuge at 4°C, 10000rpm for 10min , collect the thalline, wash the thalline twice with physiological saline, and collect the wet thalline (ie, whole cells of the recombinant bacteria).
[0064] (R)-3,5-bistrifluoromethylphenethanol is prepared by using wet bacteria as the enzyme source for transformation and using 3,5-bistrifluoromethylacetophenone as the substrate for the transformation reaction. Tr...
PUM
| Property | Measurement | Unit |
|---|---|---|
| optical purity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com